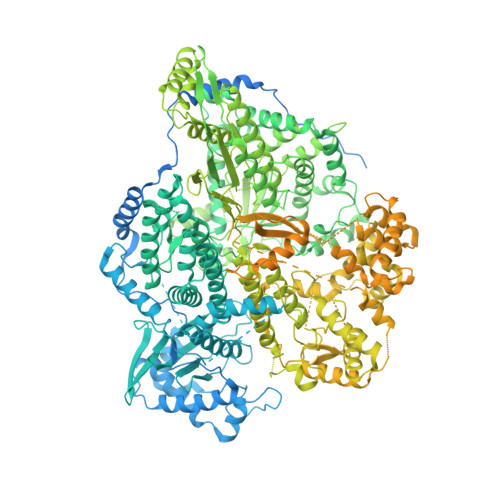
Structural and functional characterization of the Sin Nombre virus L protein.
Meier, K., Thorkelsson, S.R., Durieux Trouilleton, Q., Vogel, D., Yu, D., Kosinski, J., Cusack, S., Malet, H., Grunewald, K., Quemin, E.R.J., Rosenthal, M.(2023) PLoS Pathog 19: e1011533-e1011533
- PubMed: 37549153
- DOI: https://doi.org/10.1371/journal.ppat.1011533
- Primary Citation of Related Structures:
8CI5 - PubMed Abstract:
The Bunyavirales order is a large and diverse group of segmented negative-strand RNA viruses. Several virus families within this order contain important human pathogens, including Sin Nombre virus (SNV) of the Hantaviridae. Despite the high epidemic potential of bunyaviruses, specific medical countermeasures such as vaccines or antivirals are missing. The multifunctional ~250 kDa L protein of hantaviruses, amongst other functional domains, harbors the RNA-dependent RNA polymerase (RdRp) and an endonuclease and catalyzes transcription as well as replication of the viral RNA genome, making it a promising therapeutic target. The development of inhibitors targeting these key processes requires a profound understanding of the catalytic mechanisms. Here, we established expression and purification protocols of the full-length SNV L protein bearing the endonuclease mutation K124A. We applied different biochemical in vitro assays to provide an extensive characterization of the different enzymatic functions as well as the capacity of the hantavirus L protein to interact with the viral RNA. By using single-particle cryo-EM, we obtained a 3D model including the L protein core region containing the RdRp, in complex with the 5' promoter RNA. This first high-resolution model of a New World hantavirus L protein shows striking similarity to related bunyavirus L proteins. The interaction of the L protein with the 5' RNA observed in the structural model confirms our hypothesis of protein-RNA binding based on our biochemical data. Taken together, this study provides an excellent basis for future structural and functional studies on the hantavirus L protein and for the development of antiviral compounds.
- Bernhard Nocht Institute for Tropical Medicine (BNITM), Hamburg, Germany.
Organizational Affiliation: