Inhibitors of the Elastase LasB for the Treatment of Pseudomonas aeruginosa Lung Infections.
Konstantinovic, J., Kany, A.M., Alhayek, A., Abdelsamie, A.S., Sikandar, A., Voos, K., Yao, Y., Andreas, A., Shafiei, R., Loretz, B., Schonauer, E., Bals, R., Brandstetter, H., Hartmann, R.W., Ducho, C., Lehr, C.M., Beisswenger, C., Muller, R., Rox, K., Haupenthal, J., Hirsch, A.K.H.(2023) ACS Cent Sci 9: 2205-2215
- PubMed: 38161367
- DOI: https://doi.org/10.1021/acscentsci.3c01102
- Primary Citation of Related Structures:
8CC4 - PubMed Abstract:
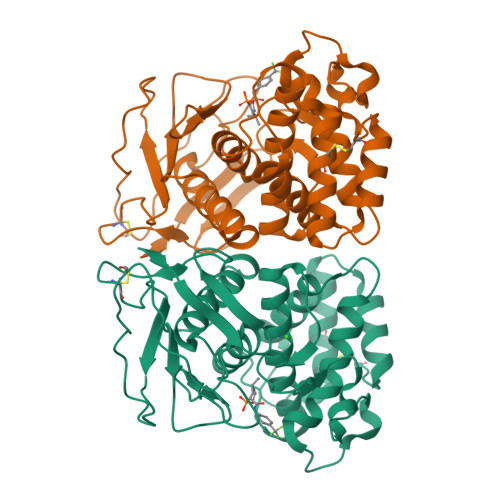
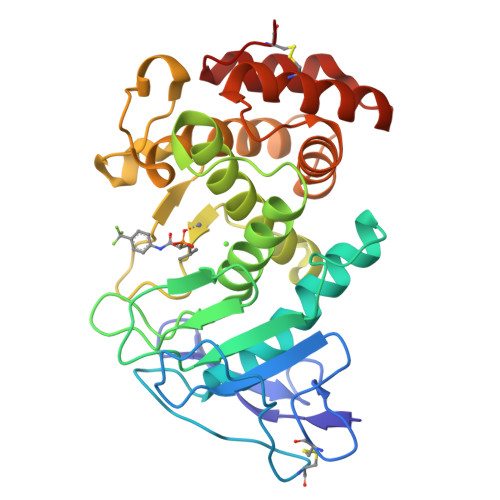
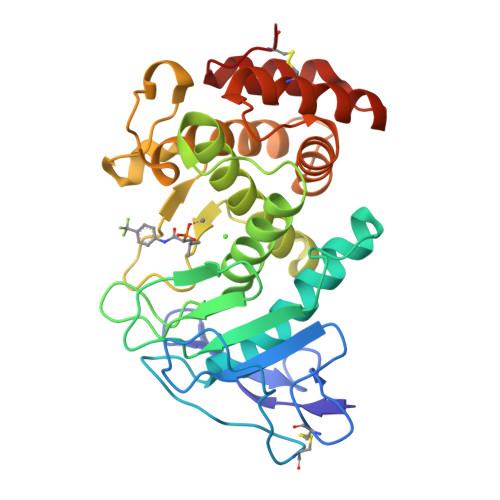
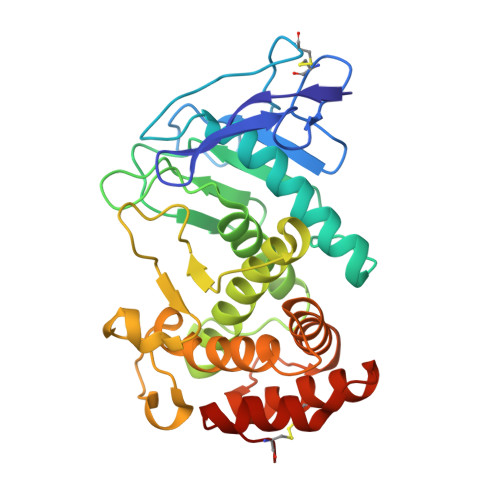
Infections caused by the Gram-negative pathogen Pseudomonas aeruginosa are emerging worldwide as a major threat to human health. Conventional antibiotic monotherapy suffers from rapid resistance development, underlining urgent need for novel treatment concepts. Here, we report on a nontraditional approach to combat P. aeruginosa -derived infections by targeting its main virulence factor, the elastase LasB. We discovered a new chemical class of phosphonates with an outstanding in vitro ADMET and PK profile, auspicious activity both in vitro and in vivo . We established the mode of action through a cocrystal structure of our lead compound with LasB and in several in vitro and ex vivo models. The proof of concept of a combination of our pathoblocker with levofloxacin in a murine neutropenic lung infection model and the reduction of LasB protein levels in blood as a proof of target engagement demonstrate the great potential for use as an adjunctive treatment of lung infections in humans.
Organizational Affiliation:
Helmholtz Institute for Pharmaceutical Research Saarland (HIPS)-Helmholtz Centre for Infection Research (HZI), Saarbrücken 66123, Germany.