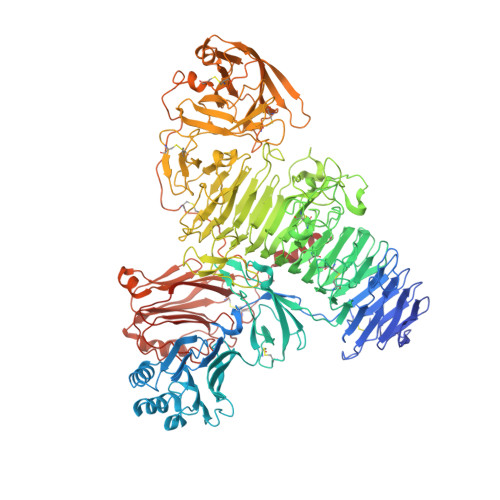
Structure of the transmembrane protein 2 (TMEM2) ectodomain and its apparent lack of hyaluronidase activity.
Niu, M., McGrath, M., Sammon, D., Gardner, S., Morgan, R.M., Di Maio, A., Liu, Y., Bubeck, D., Hohenester, E.(2023) Wellcome Open Res 8: 76-76
- PubMed: 37234743
- DOI: https://doi.org/10.12688/wellcomeopenres.18937.2
- Primary Citation of Related Structures:
8C6I - PubMed Abstract:
Background: Hyaluronic acid (HA) is a major polysaccharide component of the extracellular matrix. HA has essential functions in tissue architecture and the regulation of cell behaviour. HA turnover needs to be finely balanced. Increased HA degradation is associated with cancer, inflammation, and other pathological situations. Transmembrane protein 2 (TMEM2) is a cell surface protein that has been reported to degrade HA into ~5 kDa fragments and play an essential role in systemic HA turnover. Methods: We produced the soluble TMEM2 ectodomain (residues 106-1383; sTMEM2) in human embryonic kidney cells (HEK293) and determined its structure using X-ray crystallography. We tested sTMEM2 hyaluronidase activity using fluorescently labelled HA and size fractionation of reaction products. We tested HA binding in solution and using a glycan microarray. Results: Our crystal structure of sTMEM2 confirms a remarkably accurate prediction by AlphaFold. sTMEM2 contains a parallel β-helix typical of other polysaccharide-degrading enzymes, but an active site cannot be assigned with confidence. A lectin-like domain is inserted into the β-helix and predicted to be functional in carbohydrate binding. A second lectin-like domain at the C-terminus is unlikely to bind carbohydrates. We did not observe HA binding in two assay formats, suggesting a modest affinity at best. Unexpectedly, we were unable to observe any HA degradation by sTMEM2. Our negative results set an upper limit for k cat of approximately 10 -5 min -1 . Conclusions: Although sTMEM2 contains domain types consistent with its suggested role in TMEM2 degradation, its hyaluronidase activity was undetectable. HA degradation by TMEM2 may require additional proteins and/or localisation at the cell surface.
- Department of Life Sciences, Imperial College London, London, SW7 2AZ, UK.
Organizational Affiliation: