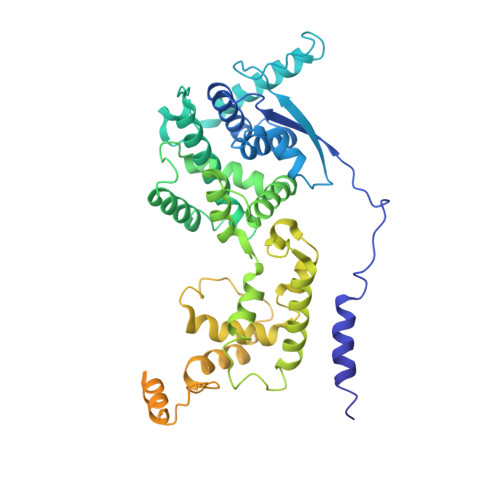
The cryoEM structure of the Hendra henipavirus nucleoprotein reveals insights into paramyxoviral nucleocapsid architectures.
Passchier, T.C., White, J.B.R., Maskell, D.P., Byrne, M.J., Ranson, N.A., Edwards, T.A., Barr, J.N.(2024) Sci Rep 14: 14099-14099
- PubMed: 38890308
- DOI: https://doi.org/10.1038/s41598-024-58243-z
- Primary Citation of Related Structures:
8C4H, 8CBW - PubMed Abstract:
We report the first cryoEM structure of the Hendra henipavirus nucleoprotein in complex with RNA, at 3.5 Å resolution, derived from single particle analysis of a double homotetradecameric RNA-bound N protein ring assembly exhibiting D14 symmetry. The structure of the HeV N protein adopts the common bi-lobed paramyxoviral N protein fold; the N-terminal and C-terminal globular domains are bisected by an RNA binding cleft containing six RNA nucleotides and are flanked by the N-terminal and C-terminal arms, respectively. In common with other paramyxoviral nucleocapsids, the lateral interface between adjacent N i and N i+1 protomers involves electrostatic and hydrophobic interactions mediated primarily through the N-terminal arm and globular domains with minor contribution from the C-terminal arm. However, the HeV N multimeric assembly uniquely identifies an additional protomer-protomer contact between the N i+1 N-terminus and N i-1 C-terminal arm linker. The model presented here broadens the understanding of RNA-bound paramyxoviral nucleocapsid architectures and provides a platform for further insight into the molecular biology of HeV, as well as the development of antiviral interventions.
- Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, LS2 9JT, UK. tim.passchier@york.ac.uk.
Organizational Affiliation: