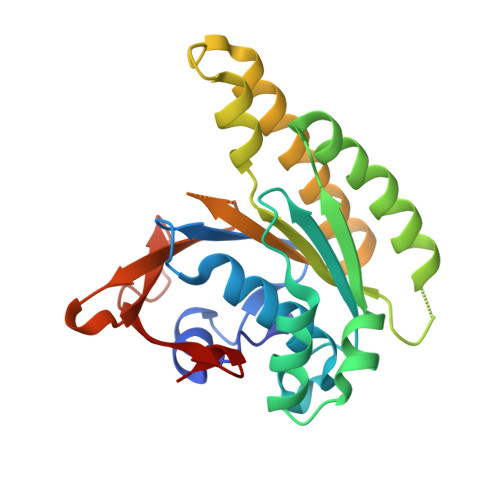
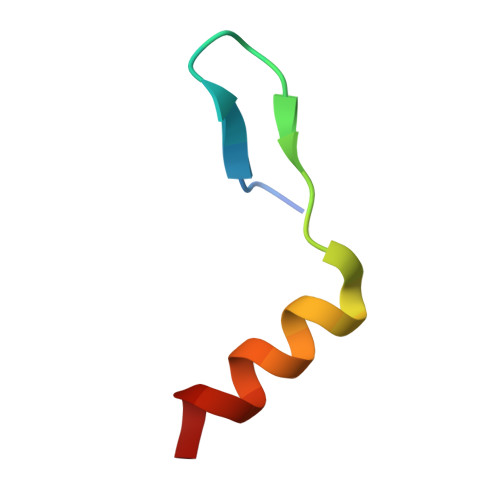
A recombinant approach for stapled peptide discovery yields inhibitors of the RAD51 recombinase.
Pantelejevs, T., Zuazua-Villar, P., Koczy, O., Counsell, A.J., Walsh, S.J., Robertson, N.S., Spring, D.R., Downs, J.A., Hyvonen, M.(2023) Chem Sci 14: 13915-13923
- PubMed: 38075664
- DOI: https://doi.org/10.1039/d3sc03331g
- Primary Citation of Related Structures:
8BR9, 8C3J, 8C3N - PubMed Abstract:
Stapling is a macrocyclisation method that connects amino acid side chains of a peptide to improve its pharmacological properties. We describe an approach for stapled peptide preparation and biochemical evaluation that combines recombinant expression of fusion constructs of target peptides and cysteine-reactive divinyl-heteroaryl chemistry as an alternative to solid-phase synthesis. We then employ this workflow to prepare and evaluate BRC-repeat-derived inhibitors of the RAD51 recombinase, showing that a diverse range of secondary structure elements in the BRC repeat can be stapled without compromising binding and function. Using X-ray crystallography, we elucidate the atomic-level features of the staple moieties. We then demonstrate that BRC-repeat-derived stapled peptides can disrupt RAD51 function in cells following ionising radiation treatment.
- Department of Biochemistry, University of Cambridge CB2 1GA UK teodors.pantelejevs@osi.lv mh256@cam.ac.uk.
Organizational Affiliation: