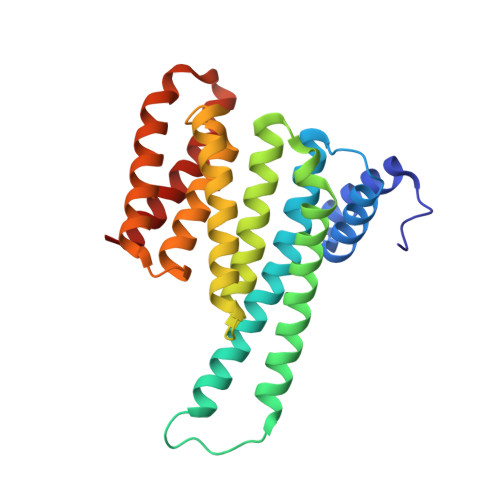
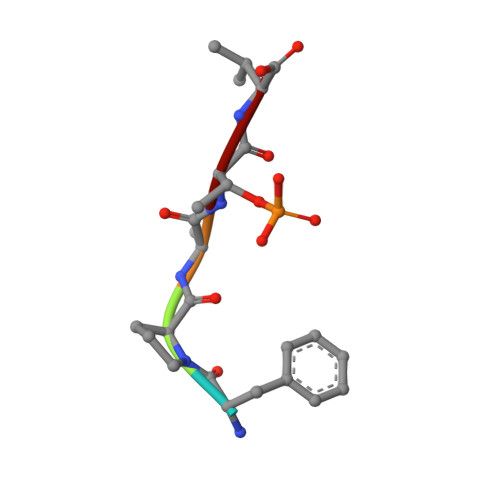
Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a new potential intervention strategy for endocrine resistance in breast cancer
Visser, E.J., Donaldson Collier, M., Siefert, J., Konstantinidou, M., Paul, S., Cossar, P., Miley, G., Meijer, O., Arkin, M.R., Ottmann, C., Zwart, W., Brunsveld, L.To be published.