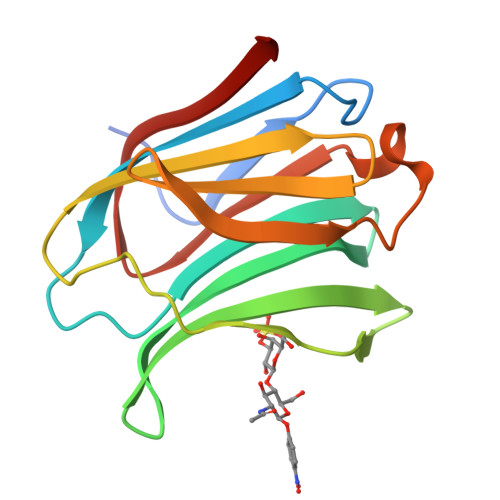
Selectively Modified Lactose and N -Acetyllactosamine Analogs at Three Key Positions to Afford Effective Galectin-3 Ligands.
Abdullayev, S., Kadav, P., Bandyopadhyay, P., Medrano, F.J., Rabinovich, G.A., Dam, T.K., Romero, A., Roy, R.(2023) Int J Mol Sci 24
- PubMed: 36835132
- DOI: https://doi.org/10.3390/ijms24043718
- Primary Citation of Related Structures:
8BZ3 - PubMed Abstract:
Galectins constitute a family of galactose-binding lectins overly expressed in the tumor microenvironment as well as in innate and adaptive immune cells, in inflammatory diseases. Lactose ((β-D-galactopyranosyl)-(1→4)-β-D-glucopyranose, Lac) and N -Acetyllactosamine (2-acetamido-2-deoxy-4- O -β-D-galactopyranosyl-D-glucopyranose, LacNAc) have been widely exploited as ligands for a wide range of galectins, sometimes with modest selectivity. Even though several chemical modifications at single positions of the sugar rings have been applied to these ligands, very few examples combined the simultaneous modifications at key positions known to increase both affinity and selectivity. We report herein combined modifications at the anomeric position, C-2, and O -3' of each of the two sugars, resulting in a 3'- O -sulfated LacNAc analog having a Kd of 14.7 µM against human Gal-3 as measured by isothermal titration calorimetry (ITC). This represents a six-fold increase in affinity when compared to methyl β-D-lactoside having a Kd of 91 µM. The three best compounds contained sulfate groups at the O -3' position of the galactoside moieties, which were perfectly in line with the observed highly cationic character of the human Gal-3 binding site shown by the co-crystal of one of the best candidates of the LacNAc series.
Organizational Affiliation:
Glycosciences and Nanomaterials Laboratory, Université du Québec à Montréal, Succ. Centre-Ville, P.O. Box 8888, Montréal, QC H3C 3P8, Canada.