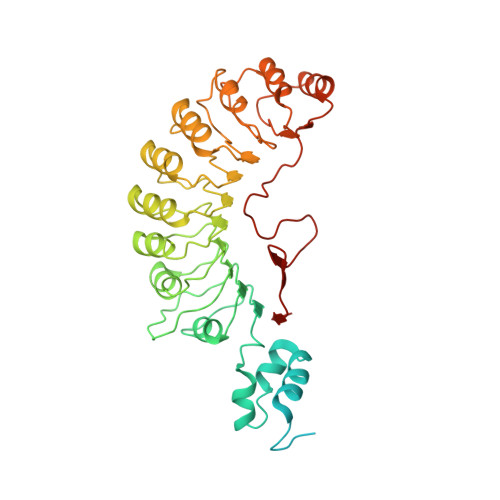
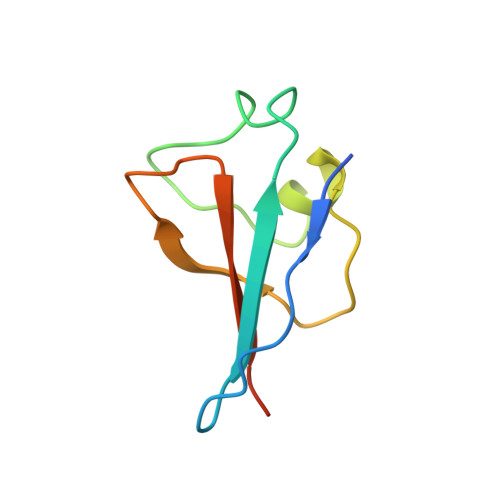
Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Rowland, R.J., Heath, R., Maskell, D., Thompson, R.F., Ranson, N.A., Blaza, J.N., Endicott, J.A., Noble, M.E.M., Salamina, M.(2023) Sci Rep 13: 10718-10718
- PubMed: 37400515
- DOI: https://doi.org/10.1038/s41598-023-37609-9
- Primary Citation of Related Structures:
8BYA, 8BYL, 8BZO - PubMed Abstract:
p27KIP1 (cyclin-dependent kinase inhibitor 1B, p27) is a member of the CIP/KIP family of CDK (cyclin dependent kinase) regulators that inhibit cell cycle CDKs. p27 phosphorylation by CDK1/2, signals its recruitment to the SCF SKP2 (S-phase kinase associated protein 1 (SKP1)-cullin-SKP2) E3 ubiquitin ligase complex for proteasomal degradation. The nature of p27 binding to SKP2 and CKS1 was revealed by the SKP1-SKP2-CKS1-p27 phosphopeptide crystal structure. Subsequently, a model for the hexameric CDK2-cyclin A-CKS1-p27-SKP1-SKP2 complex was proposed by overlaying an independently determined CDK2-cyclin A-p27 structure. Here we describe the experimentally determined structure of the isolated CDK2-cyclin A-CKS1-p27-SKP1-SKP2 complex at 3.4 Å global resolution using cryogenic electron microscopy. This structure supports previous analysis in which p27 was found to be structurally dynamic, transitioning from disordered to nascent secondary structure on target binding. We employed 3D variability analysis to further explore the conformational space of the hexameric complex and uncovered a previously unidentified hinge motion centred on CKS1. This flexibility gives rise to open and closed conformations of the hexameric complex that we propose may contribute to p27 regulation by facilitating recognition with SCF SKP2 . This 3D variability analysis further informed particle subtraction and local refinement approaches to enhance the local resolution of the complex.
- Translational and Clinical Research Institute, Newcastle University Centre for Cancer, Newcastle University, Paul O'Gorman Building, Framlington Place, Newcastle Upon Tyne, NE2 4HH, UK.
Organizational Affiliation: