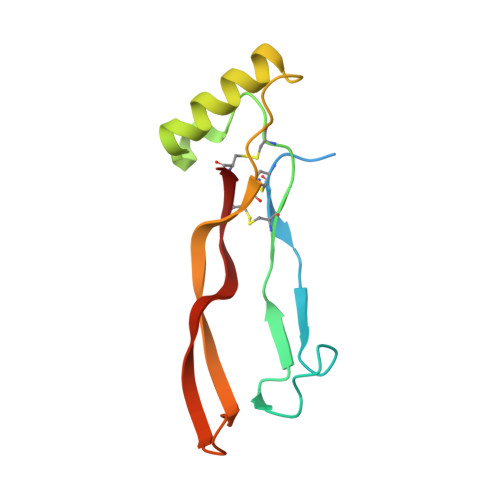
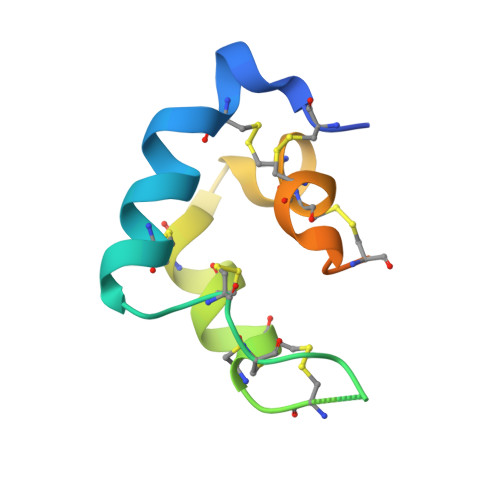
Molecular mechanism of BMP signal control by Twisted gastrulation.
Malinauskas, T., Moore, G., Rudolf, A.F., Eggington, H., Belnoue-Davis, H.L., El Omari, K., Griffiths, S.C., Woolley, R.E., Duman, R., Wagner, A., Leedham, S.J., Baldock, C., Ashe, H.L., Siebold, C.(2024) Nat Commun 15: 4976-4976
- PubMed: 38862520
- DOI: https://doi.org/10.1038/s41467-024-49065-8
- Primary Citation of Related Structures:
8BWA, 8BWD, 8BWI, 8BWL, 8BWM, 8BWN - PubMed Abstract:
Twisted gastrulation (TWSG1) is an evolutionarily conserved secreted glycoprotein which controls signaling by Bone Morphogenetic Proteins (BMPs). TWSG1 binds BMPs and their antagonist Chordin to control BMP signaling during embryonic development, kidney regeneration and cancer. We report crystal structures of TWSG1 alone and in complex with a BMP ligand, Growth Differentiation Factor 5. TWSG1 is composed of two distinct, disulfide-rich domains. The TWSG1 N-terminal domain occupies the BMP type 1 receptor binding site on BMPs, whereas the C-terminal domain binds to a Chordin family member. We show that TWSG1 inhibits BMP function in cellular signaling assays and mouse colon organoids. This inhibitory function is abolished in a TWSG1 mutant that cannot bind BMPs. The same mutation in the Drosophila TWSG1 ortholog Tsg fails to mediate BMP gradient formation required for dorsal-ventral axis patterning of the early embryo. Our studies reveal the evolutionarily conserved mechanism of BMP signaling inhibition by TWSG1.
- Division of Structural Biology, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Oxford, OX3 7BN, UK. tomas@strubi.ox.ac.uk.
Organizational Affiliation: