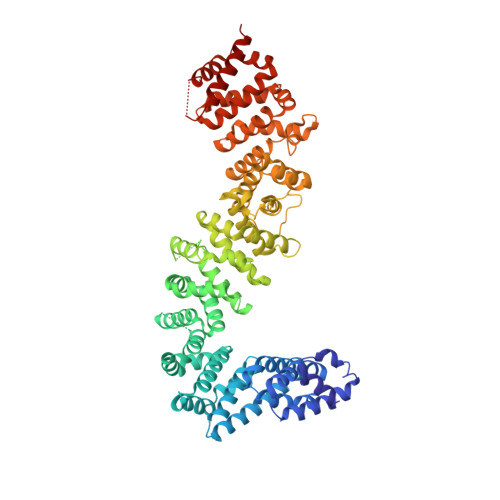
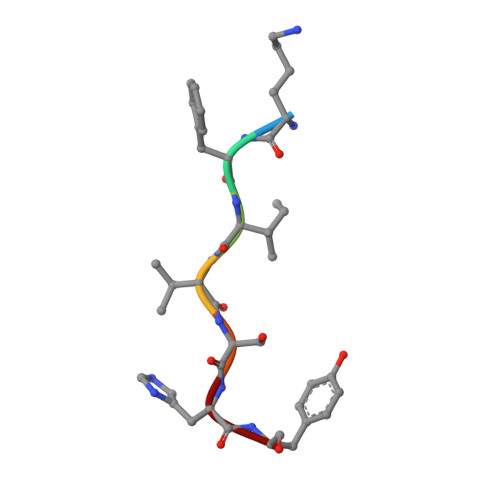
UNC-45 assisted myosin folding depends on a conserved FX 3 HY motif implicated in Freeman Sheldon Syndrome.
Vogel, A., Arnese, R., Gudino Carrillo, R.M., Sehr, D., Deszcz, L., Bylicki, A., Meinhart, A., Clausen, T.(2024) Nat Commun 15: 6272-6272
- PubMed: 39054317
- DOI: https://doi.org/10.1038/s41467-024-50442-6
- Primary Citation of Related Structures:
8BRG, 8BRH - PubMed Abstract:
Myosin motors are critical for diverse motility functions, ranging from cytokinesis and endocytosis to muscle contraction. The UNC-45 chaperone controls myosin function mediating the folding, assembly, and degradation of the muscle protein. Here, we analyze the molecular mechanism of UNC-45 as a hub in myosin quality control. We show that UNC-45 forms discrete complexes with folded and unfolded myosin, forwarding them to downstream chaperones and E3 ligases. Structural analysis of a minimal chaperone:substrate complex reveals that UNC-45 binds to a conserved FX 3 HY motif in the myosin motor domain. Disrupting the observed interface by mutagenesis prevents myosin maturation leading to protein aggregation in vivo. We also show that a mutation in the FX 3 HY motif linked to the Freeman Sheldon Syndrome impairs UNC-45 assisted folding, reducing the level of functional myosin. These findings demonstrate that a faulty myosin quality control is a critical yet unexplored cause of human myopathies.
- Research Institute of Molecular Pathology, Vienna BioCenter, Vienna, Austria.
Organizational Affiliation: