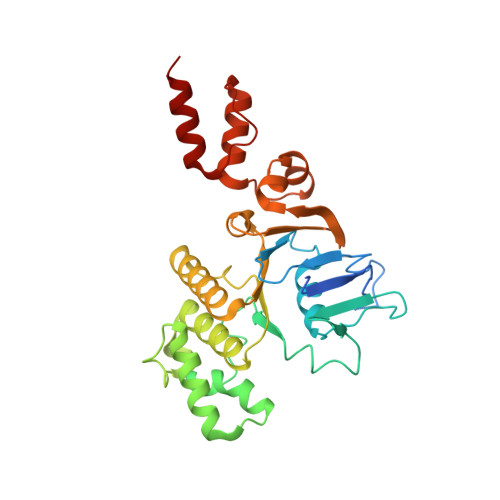
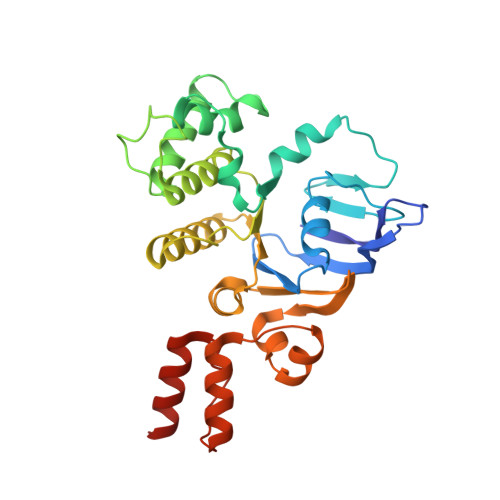
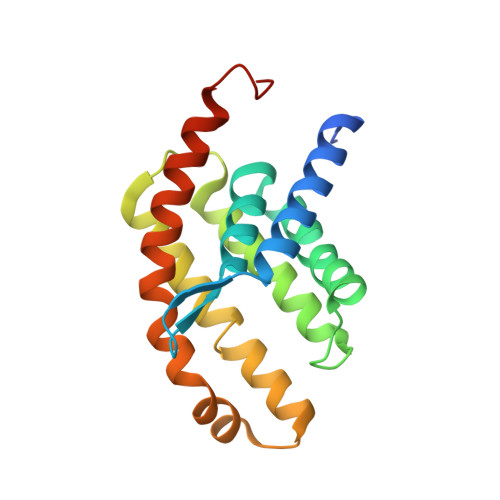
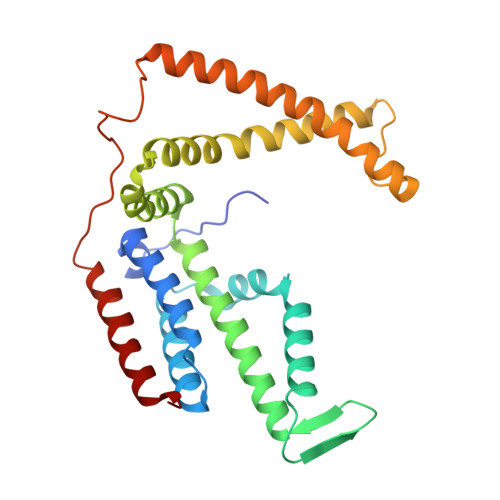
Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Thangaratnarajah, C., Nijland, M., Borges-Araujo, L., Jeucken, A., Rheinberger, J., Marrink, S.J., Souza, P.C.T., Paulino, C., Slotboom, D.J.(2023) Nat Commun 14: 4484-4484
- PubMed: 37491368
- DOI: https://doi.org/10.1038/s41467-023-40266-1
- Primary Citation of Related Structures:
8BMP, 8BMQ, 8BMR, 8BMS - PubMed Abstract:
Energy-coupling factor (ECF)-type transporters mediate the uptake of micronutrients in many bacteria. They consist of a substrate-translocating subunit (S-component) and an ATP-hydrolysing motor (ECF module) Previous data indicate that the S-component topples within the membrane to alternately expose the binding site to either side of the membrane. In many ECF transporters, the substrate-free S-component can be expelled from the ECF module. Here we study this enigmatic expulsion step by cryogenic electron microscopy and reveal that ATP induces a concave-to-convex shape change of two long helices in the motor, thereby destroying the S-component's docking site and allowing for its dissociation. We show that adaptation of the membrane morphology to the conformational state of the motor may favour expulsion of the substrate-free S-component when ATP is bound and docking of the substrate-loaded S-component after hydrolysis. Our work provides a picture of bilayer-assisted chemo-mechanical coupling in the transport cycle of ECF transporters.
- Faculty of Science and Engineering, Groningen Biomolecular Sciences and Biotechnology, Membrane Enzymology Group, University of Groningen, 9747 AG, Groningen, The Netherlands.
Organizational Affiliation: