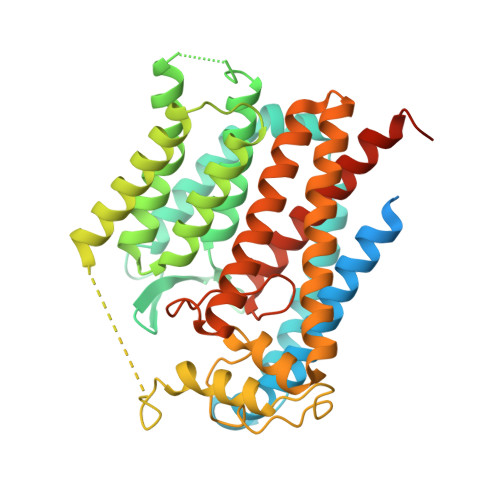
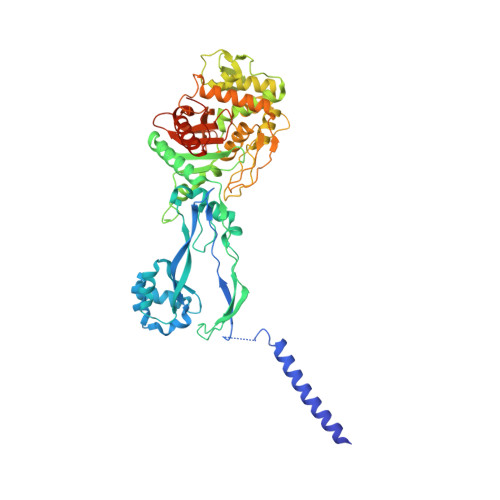
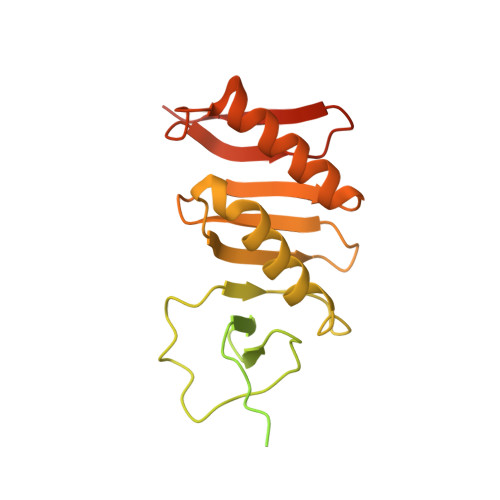
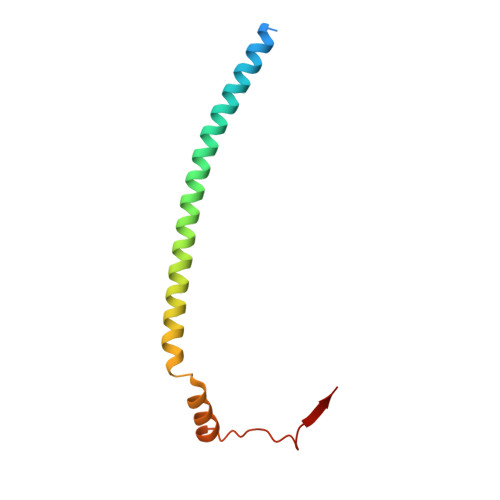
Cryo-EM structure of the bacterial divisome core complex and antibiotic target FtsWIQBL.
Kashammer, L., van den Ent, F., Jeffery, M., Jean, N.L., Hale, V.L., Lowe, J.(2023) Nat Microbiol 8: 1149-1159
- PubMed: 37127704
- DOI: https://doi.org/10.1038/s41564-023-01368-0
- Primary Citation of Related Structures:
8BH1 - PubMed Abstract:
In most bacteria, cell division relies on the synthesis of new cell wall material by the multiprotein divisome complex. Thus, at the core of the divisome are the transglycosylase FtsW, which synthesises peptidoglycan strands from its substrate Lipid II, and the transpeptidase FtsI that cross-links these strands to form a mesh, shaping and protecting the bacterial cell. The FtsQ-FtsB-FtsL trimeric complex interacts with the FtsWI complex and is involved in regulating its enzymatic activities; however, the structure of this pentameric complex is unknown. Here, we present the cryogenic electron microscopy structure of the FtsWIQBL complex from Pseudomonas aeruginosa at 3.7 Å resolution. Our work reveals intricate structural details, including an extended coiled coil formed by FtsL and FtsB and the periplasmic interaction site between FtsL and FtsI. Our structure explains the consequences of previously reported mutations and we postulate a possible activation mechanism involving a large conformational change in the periplasmic domain. As FtsWIQBL is central to the divisome, our structure is foundational for the design of future experiments elucidating the precise mechanism of bacterial cell division, an important antibiotic target.
- Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.
Organizational Affiliation: