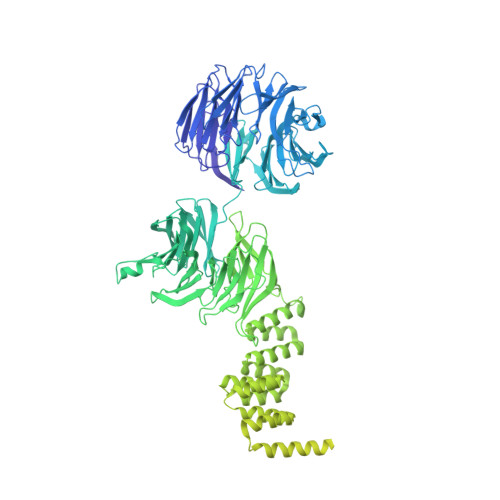
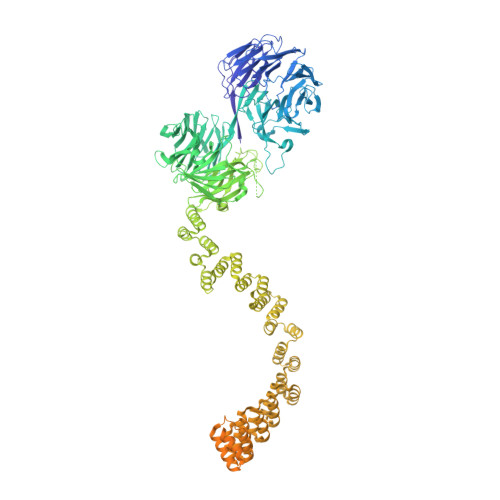
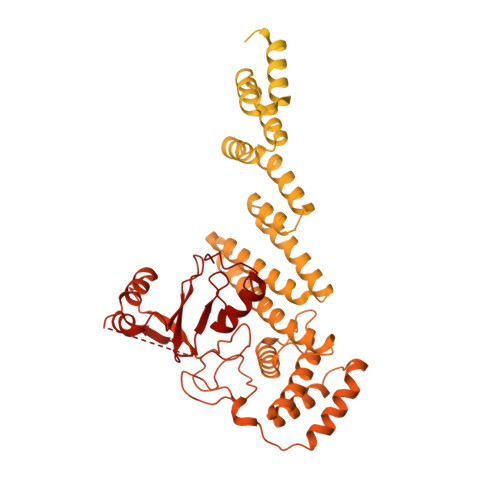
IFT-A structure reveals carriages for membrane protein transport into cilia.
Hesketh, S.J., Mukhopadhyay, A.G., Nakamura, D., Toropova, K., Roberts, A.J.(2022) Cell 185: 4971
- PubMed: 36462505
- DOI: https://doi.org/10.1016/j.cell.2022.11.010
- Primary Citation of Related Structures:
8BBE, 8BBF, 8BBG - PubMed Abstract:
Intraflagellar transport (IFT) trains are massive molecular machines that traffic proteins between cilia and the cell body. Each IFT train is a dynamic polymer of two large complexes (IFT-A and -B) and motor proteins, posing a formidable challenge to mechanistic understanding. Here, we reconstituted the complete human IFT-A complex and obtained its structure using cryo-EM. Combined with AlphaFold prediction and genome-editing studies, our results illuminate how IFT-A polymerizes, interacts with IFT-B, and uses an array of β-propeller and TPR domains to create "carriages" of the IFT train that engage TULP adaptor proteins. We show that IFT-A⋅TULP carriages are essential for cilia localization of diverse membrane proteins, as well as ICK-the key kinase regulating IFT train turnaround. These data establish a structural link between IFT-A's distinct functions, provide a blueprint for IFT-A in the train, and shed light on how IFT evolved from a proto-coatomer ancestor.
- Institute of Structural and Molecular Biology, Department of Biological Sciences, Birkbeck University of London, London, WC1E 7HX, UK.
Organizational Affiliation: