Crystal structure of NEI domain of mouse NEIL3 trapped in covalent complex with ssDNA with abasic site
Klima, M., Boura, E., Silhan, J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Endonuclease 8-like 3 | 290 | Mus musculus | Mutation(s): 0 Gene Names: Neil3 EC: 3.2.2 (PDB Primary Data), 4.2.99.18 (PDB Primary Data) | 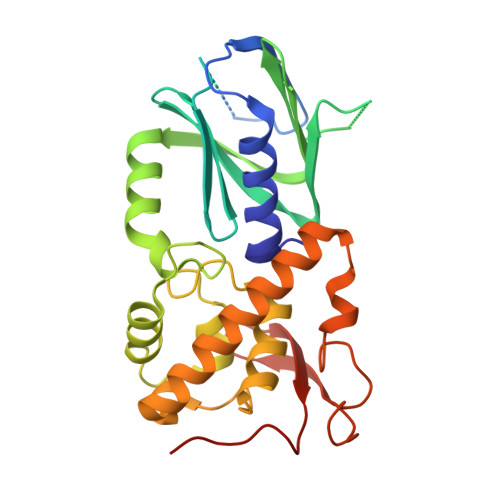 | |
UniProt | |||||
Find proteins for Q8K203 (Mus musculus) Explore Q8K203 Go to UniProtKB: Q8K203 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q8K203 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| ssDNA with abasic site | 12 | Mus musculus | 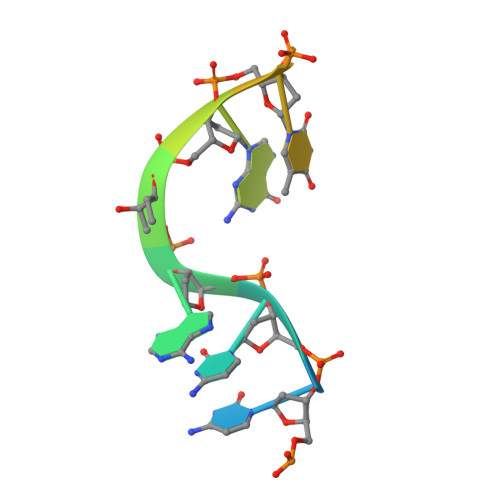 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ZN Query on ZN | E [auth A], F [auth C] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 75.056 | α = 90 |
| b = 75.056 | β = 90 |
| c = 220.292 | γ = 90 |
| Software Name | Purpose |
|---|---|
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |
| Coot | model building |
| PHENIX | refinement |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |