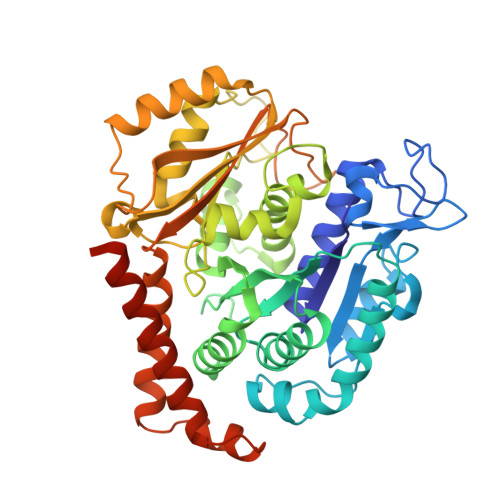
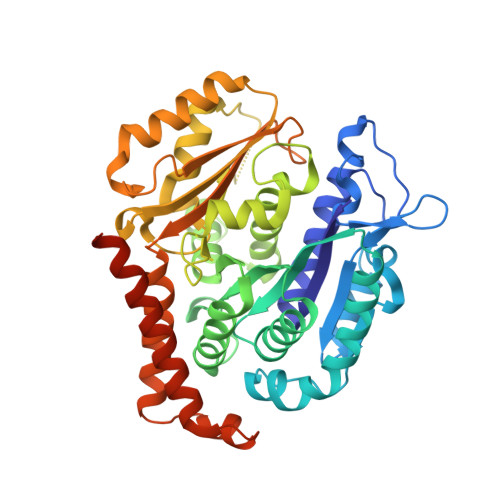
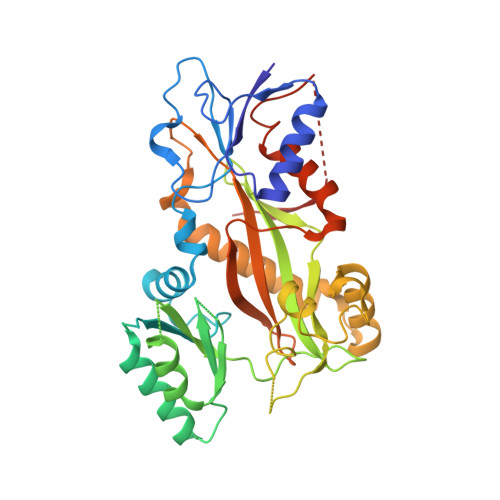
Maytansinol Functionalization: Towards Useful Probes for Studying Microtubule Dynamics.
Boiarska, Z., Perez-Pena, H., Abel, A.C., Marzullo, P., Alvarez-Bernad, B., Bonato, F., Santini, B., Horvath, D., Lucena-Agell, D., Vasile, F., Sironi, M., Diaz, J.F., Prota, A.E., Pieraccini, S., Passarella, D.(2023) Chemistry 29: e202203431-e202203431
- PubMed: 36468686
- DOI: https://doi.org/10.1002/chem.202203431
- Primary Citation of Related Structures:
8B7A, 8B7B, 8B7C - PubMed Abstract:
Maytansinoids are a successful class of natural and semisynthetic tubulin binders, known for their potent cytotoxic activity. Their wider application as cytotoxins and chemical probes to study tubulin dynamics has been held back by the complexity of natural product chemistry. Here we report the synthesis of long-chain derivatives and maytansinoid conjugates. We confirmed that bulky substituents do not impact their high activity or the scaffold's binding mode. These encouraging results open new avenues for the design of new maytansine-based probes.
- Department of Chemistry, Università degli Studi di Milano, Via Golgi 19, 20133, Milan, Italy.
Organizational Affiliation: