Structural basis for translation inhibition by the glycosylated drosocin peptide.
Koller, T.O., Morici, M., Berger, M., Safdari, H.A., Lele, D.S., Beckert, B., Kaur, K.J., Wilson, D.N.(2023) Nat Chem Biol 19: 1072-1081
- PubMed: 36997646
- DOI: https://doi.org/10.1038/s41589-023-01293-7
- Primary Citation of Related Structures:
8AKN, 8AM9, 8ANA - PubMed Abstract:
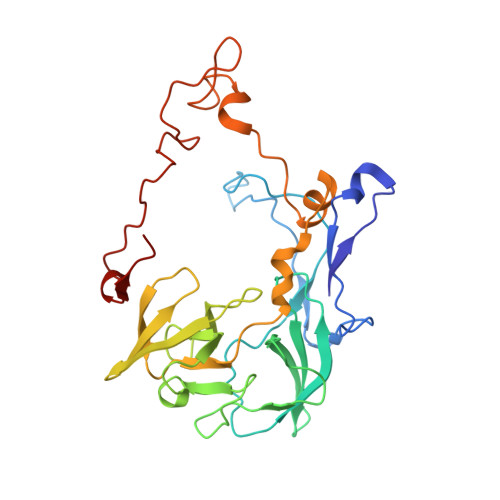
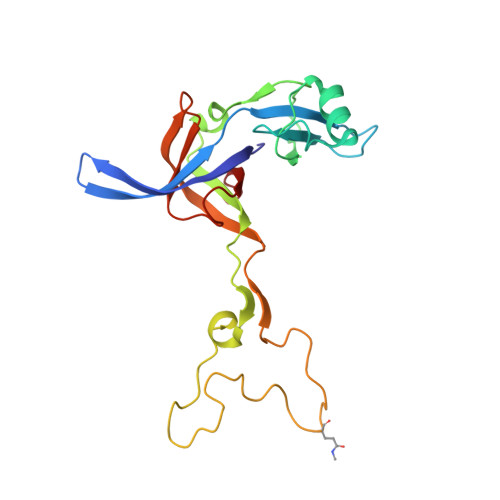
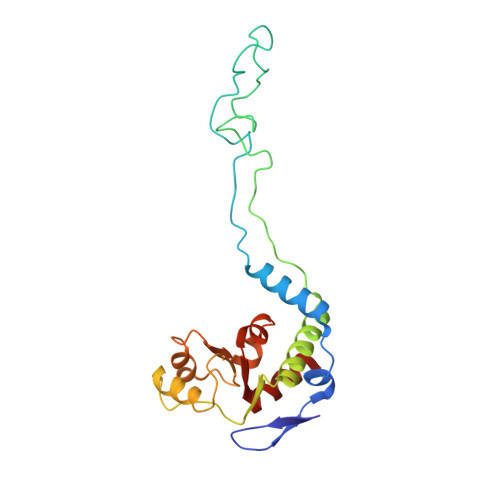
The proline-rich antimicrobial peptide (PrAMP) drosocin is produced by Drosophila species to combat bacterial infection. Unlike many PrAMPs, drosocin is O-glycosylated at threonine 11, a post-translation modification that enhances its antimicrobial activity. Here we demonstrate that the O-glycosylation not only influences cellular uptake of the peptide but also interacts with its intracellular target, the ribosome. Cryogenic electron microscopy structures of glycosylated drosocin on the ribosome at 2.0-2.8-Å resolution reveal that the peptide interferes with translation termination by binding within the polypeptide exit tunnel and trapping RF1 on the ribosome, reminiscent of that reported for the PrAMP apidaecin. The glycosylation of drosocin enables multiple interactions with U2609 of the 23S rRNA, leading to conformational changes that break the canonical base pair with A752. Collectively, our study reveals novel molecular insights into the interaction of O-glycosylated drosocin with the ribosome, which provide a structural basis for future development of this class of antimicrobials.
- Institute for Biochemistry and Molecular Biology, University of Hamburg, Hamburg, Germany.
Organizational Affiliation: