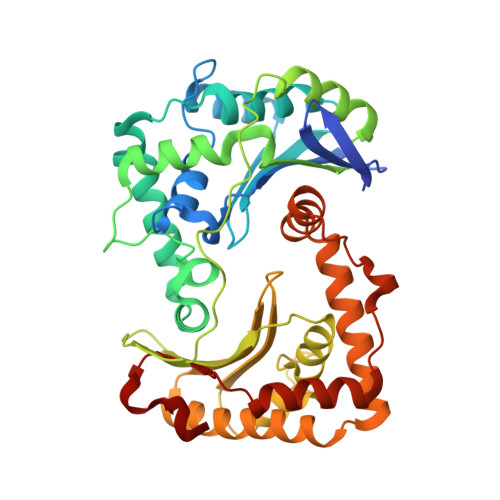
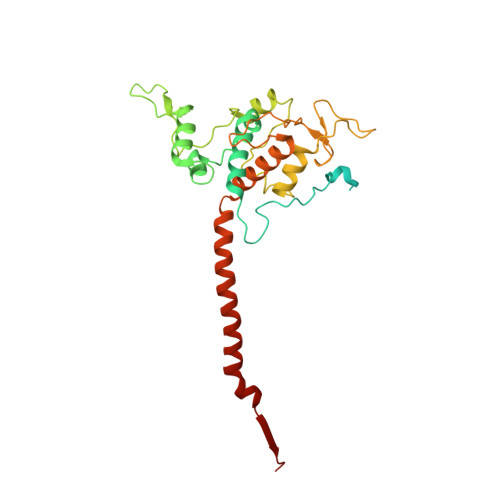
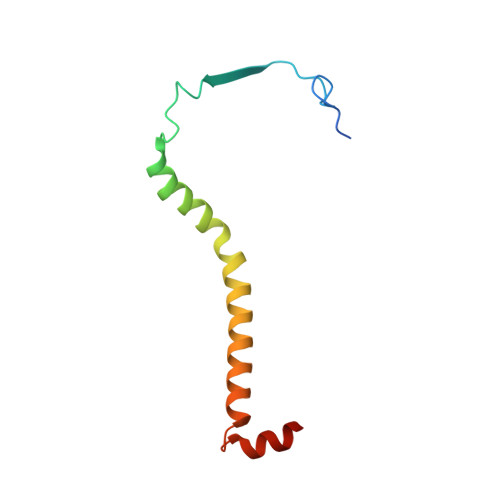
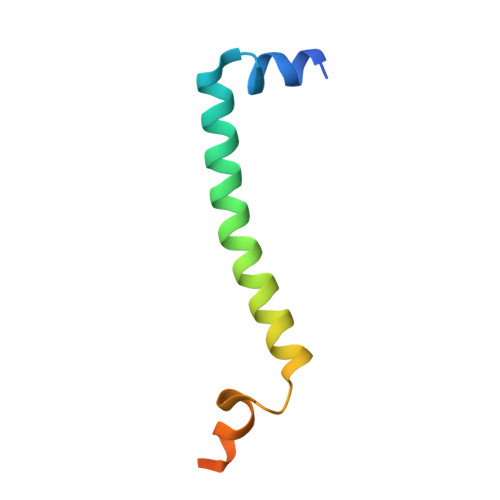
Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Wieferig, J.P., Kuhlbrandt, W.(2023) IUCrJ 10: 27-37
- PubMed: 36598500
- DOI: https://doi.org/10.1107/S2052252522010570
- Primary Citation of Related Structures:
8AB6, 8AB7, 8AB8, 8AB9, 8ABA, 8ABB, 8ABE, 8ABF, 8ABG, 8ABH, 8ABI, 8ABJ, 8ABK, 8ABL, 8ABM, 8AC3, 8AC4, 8AC5 - PubMed Abstract:
Movement of the Rieske domain of the iron-sulfur protein is essential for intramolecular electron transfer within complex III 2 (CIII 2 ) of the respiratory chain as it bridges a gap in the cofactor chain towards the electron acceptor cytochrome c. We present cryo-EM structures of CIII 2 from Yarrowia lipolytica at resolutions up to 2.0 Å under different conditions, with different redox states of the cofactors of the high-potential chain. All possible permutations of three primary positions were observed, indicating that the two halves of the dimeric complex act independently. Addition of the substrate analogue decylubiquinone to CIII 2 with a reduced high-potential chain increased the occupancy of the Q o site. The extent of Rieske domain interactions through hydrogen bonds to the cytochrome b and cytochrome c 1 subunits varied depending on the redox state and substrate. In the absence of quinols, the reduced Rieske domain interacted more closely with cytochrome b and cytochrome c 1 than in the oxidized state. Upon addition of the inhibitor antimycin A, the heterogeneity of the cd 1 -helix and ef-loop increased, which may be indicative of a long-range effect on the Rieske domain.
- Department of Structural Biology, Max Planck Institute of Biophysics, Max-von-Laue-Strasse 3, 60438 Frankfurt am Main, Germany.
Organizational Affiliation: