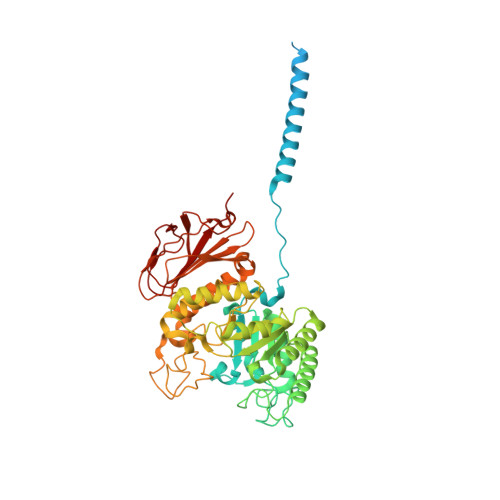
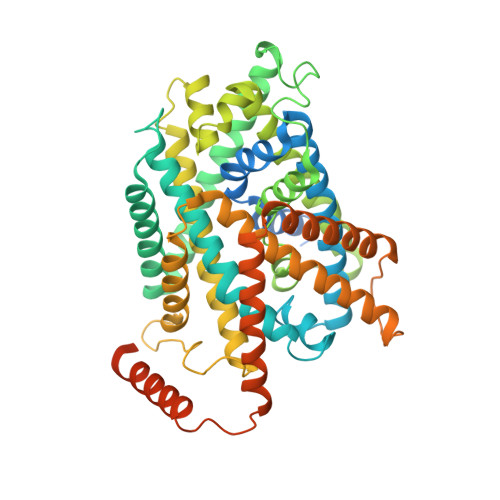
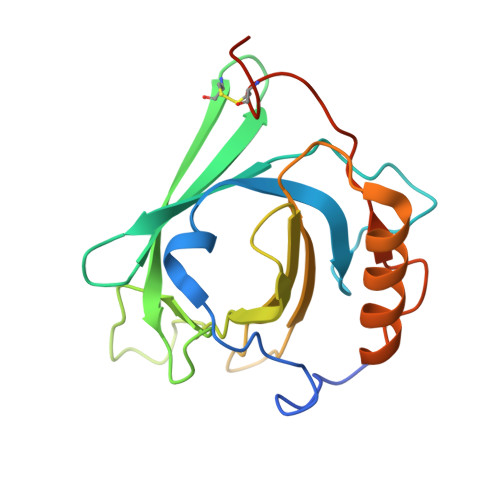
Structure of the human heterodimeric transporter 4F2hc-LAT2 in complex with Anticalin, an alternative binding protein for applications in single-particle cryo-EM.
Jeckelmann, J.M., Lemmin, T., Schlapschy, M., Skerra, A., Fotiadis, D.(2022) Sci Rep 12: 18269-18269
- PubMed: 36310334
- DOI: https://doi.org/10.1038/s41598-022-23270-1
- Primary Citation of Related Structures:
8A6L - PubMed Abstract:
Cryo-EM structure determination of relatively small and flexible membrane proteins at high resolution is challenging. Increasing the size and structural features by binding of high affinity proteins to the biomolecular target allows for better particle alignment and may result in structural models of higher resolution and quality. Anticalins are alternative binding proteins to antibodies, which are based on the lipocalin scaffold and show potential for theranostic applications. The human heterodimeric amino acid transporter 4F2hc-LAT2 is a membrane protein complex that mediates transport of certain amino acids and derivatives thereof across the plasma membrane. Here, we present and discuss the cryo-EM structure of human 4F2hc-LAT2 in complex with the anticalin D11vs at 3.2 Å resolution. Relative high local map resolution (2.8-3.0 Å) in the LAT2 substrate binding site together with molecular dynamics simulations indicated the presence of fixed water molecules potentially involved in shaping and stabilizing this region. Finally, the presented work expands the application portfolio of anticalins and widens the toolset of binding proteins to promote high-resolution structure solution by single-particle cryo-EM.
- Institute of Biochemistry and Molecular Medicine, University of Bern, Bern, Switzerland. jean-marc.jeckelmann@ibmm.unibe.ch.
Organizational Affiliation: