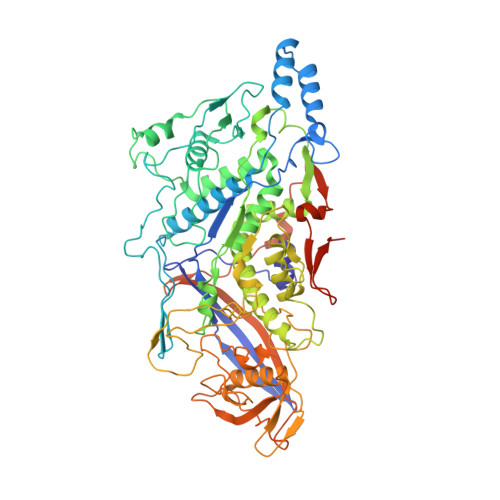
Delineating organizational principles of the endogenous L-A virus by cryo-EM and computational analysis of native cell extracts.
Schmidt, L., Tuting, C., Kyrilis, F.L., Hamdi, F., Semchonok, D.A., Hause, G., Meister, A., Ihling, C., Stubbs, M.T., Sinz, A., Kastritis, P.L.(2024) Commun Biol 7: 557-557
- PubMed: 38730276
- DOI: https://doi.org/10.1038/s42003-024-06204-7
- Primary Citation of Related Structures:
8A5T, 8PE4 - PubMed Abstract:
The high abundance of most viruses in infected host cells benefits their structural characterization. However, endogenous viruses are present in low copy numbers and are therefore challenging to investigate. Here, we retrieve cell extracts enriched with an endogenous virus, the yeast L-A virus. The determined cryo-EM structure discloses capsid-stabilizing cation-π stacking, widespread across viruses and within the Totiviridae, and an interplay of non-covalent interactions from ten distinct capsomere interfaces. The capsid-embedded mRNA decapping active site trench is supported by a constricting movement of two flexible opposite-facing loops. tRNA-loaded polysomes and other biomacromolecules, presumably mRNA, are found in virus proximity within the cell extract. Mature viruses participate in larger viral communities resembling their rare in-cell equivalents in terms of size, composition, and inter-virus distances. Our results collectively describe a 3D-architecture of a viral milieu, opening the door to cell-extract-based high-resolution structural virology.
- Interdisciplinary Research Center HALOmem, Charles Tanford Protein Center, Martin Luther University Halle-Wittenberg, Kurt-Mothes-Straße 3a, Halle/Saale, Germany.
Organizational Affiliation: