Chemical proteomics reveals the target landscape of 1,000 kinase inhibitors.
Reinecke, M., Brear, P., Vornholz, L., Berger, B.T., Seefried, F., Wilhelm, S., Samaras, P., Gyenis, L., Litchfield, D.W., Medard, G., Muller, S., Ruland, J., Hyvonen, M., Wilhelm, M., Kuster, B.(2024) Nat Chem Biol 20: 577-585
- PubMed: 37904048
- DOI: https://doi.org/10.1038/s41589-023-01459-3
- Primary Citation of Related Structures:
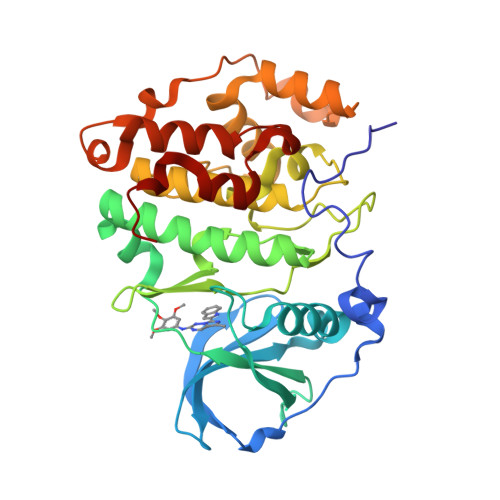
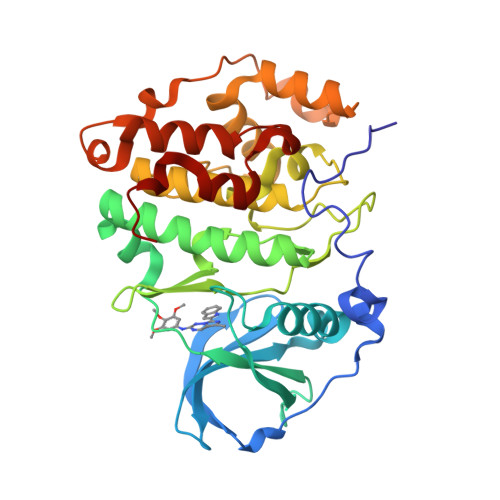
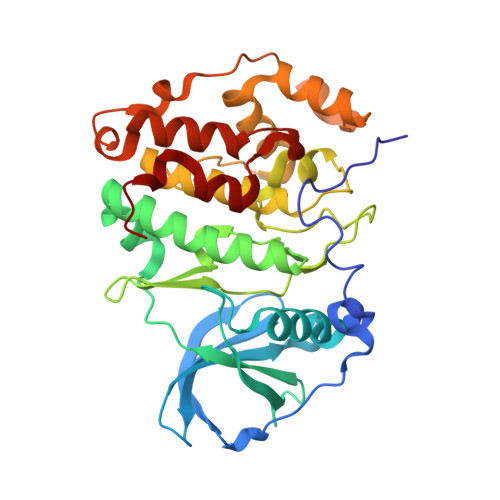
7A4Q, 7ZWE, 7ZWG - PubMed Abstract:
Medicinal chemistry has discovered thousands of potent protein and lipid kinase inhibitors. These may be developed into therapeutic drugs or chemical probes to study kinase biology. Because of polypharmacology, a large part of the human kinome currently lacks selective chemical probes. To discover such probes, we profiled 1,183 compounds from drug discovery projects in lysates of cancer cell lines using Kinobeads. The resulting 500,000 compound-target interactions are available in ProteomicsDB and we exemplify how this molecular resource may be used. For instance, the data revealed several hundred reasonably selective compounds for 72 kinases. Cellular assays validated GSK986310C as a candidate SYK (spleen tyrosine kinase) probe and X-ray crystallography uncovered the structural basis for the observed selectivity of the CK2 inhibitor GW869516X. Compounds targeting PKN3 were discovered and phosphoproteomics identified substrates that indicate target engagement in cells. We anticipate that this molecular resource will aid research in drug discovery and chemical biology.
Organizational Affiliation:
Chair of Proteomics and Bioanalytics, Technical University of Munich, Freising, Germany.