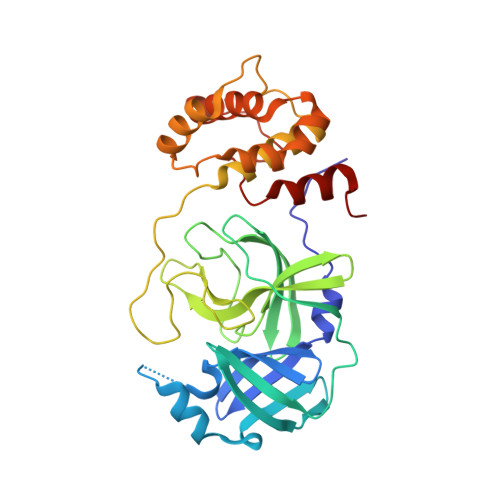
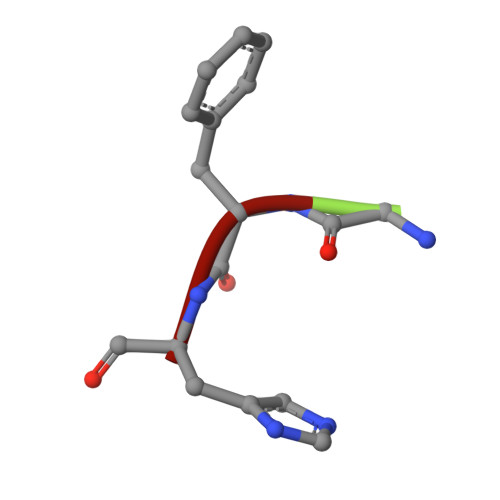
Rational design of the zonulin inhibitor AT1001 derivatives as potential anti SARS-CoV-2.
Di Micco, S., Rahimova, R., Sala, M., Scala, M.C., Vivenzio, G., Musella, S., Andrei, G., Remans, K., Mammri, L., Snoeck, R., Bifulco, G., Di Matteo, F., Vestuto, V., Campiglia, P., Marquez, J.A., Fasano, A.(2022) Eur J Med Chem 244: 114857-114857
- PubMed: 36332548
- DOI: https://doi.org/10.1016/j.ejmech.2022.114857
- Primary Citation of Related Structures:
7ZV5, 7ZV7, 7ZV8 - PubMed Abstract:
Although vaccines are greatly mitigating the worldwide pandemic diffusion of SARS-Cov-2, therapeutics should provide many distinct advantages as complementary approach to control the viral spreading. Here, we report the development of new tripeptide derivatives of AT1001 against SARS-CoV-2 M pro . By molecular modeling, a small compound library was rationally designed and filtered for enzymatic inhibition through FRET assay, leading to the identification of compound 4. X-ray crystallography studies provide insights into its binding mode and confirm the formation of a covalent bond with M pro C145. In vitro antiviral tests indicate the improvement of biological activity of 4 respect to AT1001. In silico and X-ray crystallography analysis led to 58, showing a promising activity against three SARS-CoV-2 variants and a valuable safety in Vero cells and human embryonic lung fibroblasts. The drug tolerance was also confirmed by in vivo studies, along with pharmacokinetics evaluation. In summary, 58 could pave the way to develop a clinical candidate for intranasal administration.
- European Biomedical Research Institute of Salerno (EBRIS), Via Salvatore de Renzi 50, 84125, Salerno, Italy. Electronic address: s.dimicco@ebris.eu.
Organizational Affiliation: