Stereoselective synthesis of 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-one derivatives as PIM kinase inhibitors inspired from marine alkaloids.
Casuscelli, F., Ardini, E., Avanzi, N., Badari, A., Casale, E., Disingrini, T., Donati, D., Ermoli, A., Felder, E.R., Galvani, A., Isacchi, A., Menichincheri, M., Montemartini, M., Orrenius, C., Piutti, C., Salom, B., Papeo, G.(2022) Chirality 34: 1437-1452
- PubMed: 35959859
- DOI: https://doi.org/10.1002/chir.23501
- Primary Citation of Related Structures:
7ZUN - PubMed Abstract:
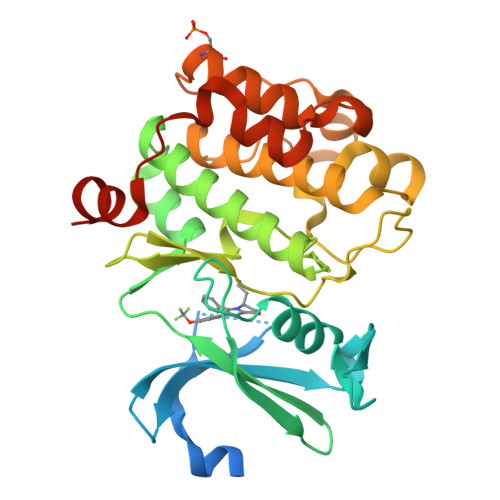
We previously demonstrated that natural product-inspired 3,4-dihydropyrrolo[1,2-a]pyrazin-1(2H)-ones derivatives delivered potent and selective PIM kinases inhibitors however with non-optimal ADME/PK properties and modest oral bioavailability. Herein, we describe a structure-based scaffold decoration and a stereoselective approach to this chemical class. The synthesis, structure-activity relationship studies, chiral analysis, and pharmacokinetic data of compounds from this inhibitor class are presented herein. Compound 20c demonstrated excellent potency on PIM1 and PIM2 with exquisite kinases selectivity and PK properties that efficiently and dose-dependently promoted c-Myc degradation and appear to be promising lead compounds for further development.
Organizational Affiliation:
Oncology, Nerviano Medical Sciences, Nerviano (Mi), Italy.