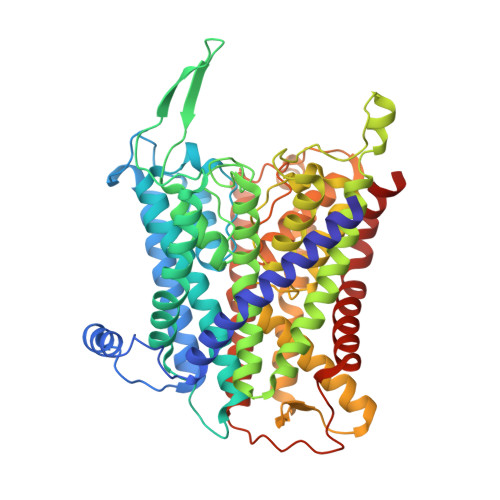
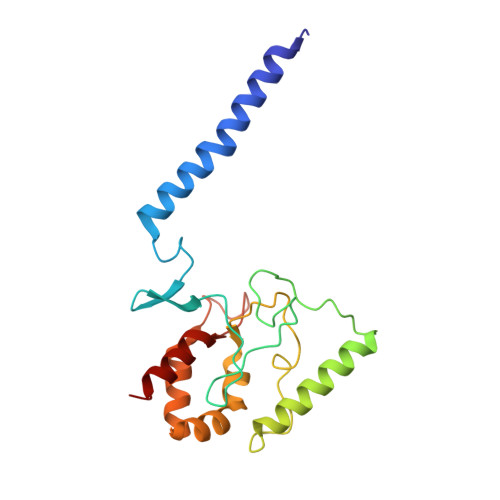
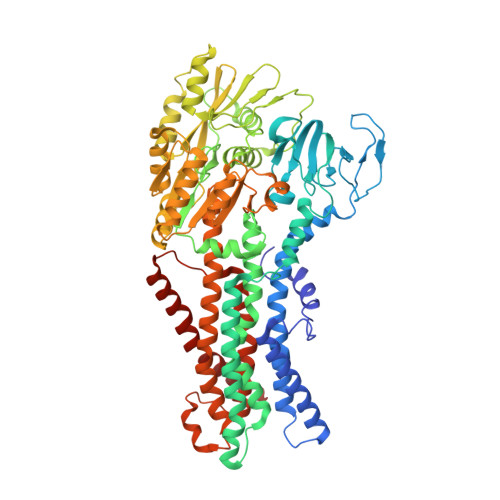
Inhibited KdpFABC transitions into an E1 off-cycle state.
Silberberg, J.M., Stock, C., Hielkema, L., Corey, R.A., Rheinberger, J., Wunnicke, D., Dubach, V.R.A., Stansfeld, P.J., Hanelt, I., Paulino, C.(2022) Elife 11
- PubMed: 36255052
- DOI: https://doi.org/10.7554/eLife.80988
- Primary Citation of Related Structures:
7ZRD, 7ZRE, 7ZRG, 7ZRH, 7ZRI, 7ZRJ, 7ZRK, 7ZRL, 7ZRM - PubMed Abstract:
KdpFABC is a high-affinity prokaryotic K + uptake system that forms a functional chimera between a channel-like subunit (KdpA) and a P-type ATPase (KdpB). At high K + levels, KdpFABC needs to be inhibited to prevent excessive K + accumulation to the point of toxicity. This is achieved by a phosphorylation of the serine residue in the TGES 162 motif in the A domain of the pump subunit KdpB (KdpB S162-P ). Here, we explore the structural basis of inhibition by KdpB S162 phosphorylation by determining the conformational landscape of KdpFABC under inhibiting and non-inhibiting conditions. Under turnover conditions, we identified a new inhibited KdpFABC state that we termed E1P tight, which is not part of the canonical Post-Albers transport cycle of P-type ATPases. It likely represents the biochemically described stalled E1P state adopted by KdpFABC upon KdpB S162 phosphorylation. The E1P tight state exhibits a compact fold of the three cytoplasmic domains and is likely adopted when the transition from high-energy E1P states to E2P states is unsuccessful. This study represents a structural characterization of a biologically relevant off-cycle state in the P-type ATPase family and supports the emerging discussion of P-type ATPase regulation by such states.
- Institute of Biochemistry, Biocenter, Goethe University Frankfurt, Frankfurt, Germany.
Organizational Affiliation: