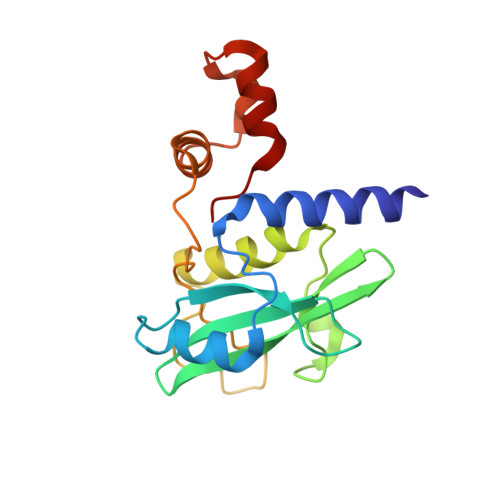
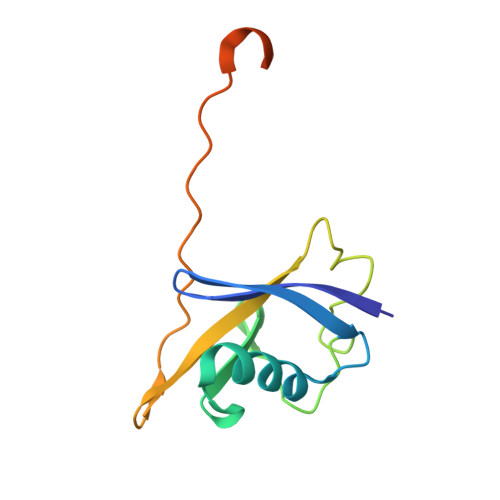
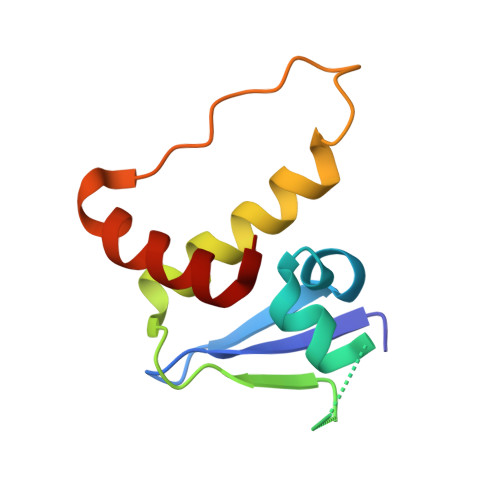
Structure-based design of a phosphotyrosine-masked covalent ligand targeting the E3 ligase SOCS2.
Ramachandran, S., Makukhin, N., Haubrich, K., Nagala, M., Forrester, B., Lynch, D.M., Casement, R., Testa, A., Bruno, E., Gitto, R., Ciulli, A.(2023) Nat Commun 14: 6345-6345
- PubMed: 37816714
- DOI: https://doi.org/10.1038/s41467-023-41894-3
- Primary Citation of Related Structures:
7ZLM, 7ZLN, 7ZLO, 7ZLP, 7ZLR, 7ZLS - PubMed Abstract:
The Src homology 2 (SH2) domain recognizes phosphotyrosine (pY) post translational modifications in partner proteins to trigger downstream signaling. Drug discovery efforts targeting the SH2 domains have long been stymied by the poor drug-like properties of phosphate and its mimetics. Here, we use structure-based design to target the SH2 domain of the E3 ligase suppressor of cytokine signaling 2 (SOCS2). Starting from the highly ligand-efficient pY amino acid, a fragment growing approach reveals covalent modification of Cys111 in a co-crystal structure, which we leverage to rationally design a cysteine-directed electrophilic covalent inhibitor MN551. We report the prodrug MN714 containing a pivaloyloxymethyl (POM) protecting group and evidence its cell permeability and capping group unmasking using cellular target engagement and in-cell 19 F NMR spectroscopy. Covalent engagement at Cys111 competitively blocks recruitment of cellular SOCS2 protein to its native substrate. The qualified inhibitors of SOCS2 could find attractive applications as chemical probes to understand the biology of SOCS2 and its CRL5 complex, and as E3 ligase handles in proteolysis targeting chimera (PROTACs) to induce targeted protein degradation.
- Centre for Targeted Protein Degradation, Division of Biological Chemistry and Drug Discovery, School of Life Sciences, University of Dundee, 1 James Lindsay Place, Dundee, DD1 5JJ, United Kingdom.
Organizational Affiliation: