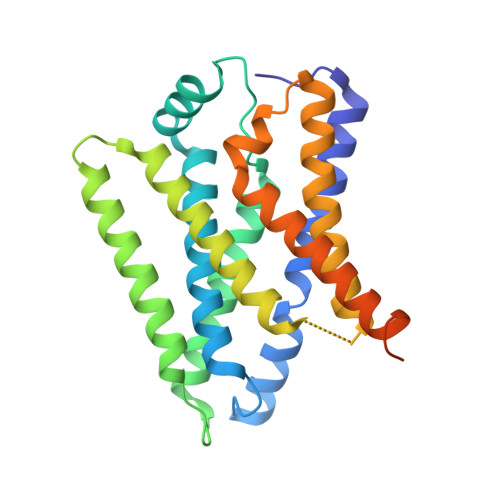
Structural basis for proton coupled cystine transport by cystinosin.
Lobel, M., Salphati, S.P., El Omari, K., Wagner, A., Tucker, S.J., Parker, J.L., Newstead, S.(2022) Nat Commun 13: 4845-4845
- PubMed: 35977944
- DOI: https://doi.org/10.1038/s41467-022-32589-2
- Primary Citation of Related Structures:
7ZK1, 7ZKW, 7ZKZ - PubMed Abstract:
Amino acid transporters play a key role controlling the flow of nutrients across the lysosomal membrane and regulating metabolism in the cell. Mutations in the gene encoding the transporter cystinosin result in cystinosis, an autosomal recessive metabolic disorder characterised by the accumulation of cystine crystals in the lysosome. Cystinosin is a member of the PQ-loop family of solute carrier (SLC) transporters and uses the proton gradient to drive cystine export into the cytoplasm. However, the molecular basis for cystinosin function remains elusive, hampering efforts to develop novel treatments for cystinosis and understand the mechanisms of ion driven transport in the PQ-loop family. To address these questions, we present the crystal structures of cystinosin from Arabidopsis thaliana in both apo and cystine bound states. Using a combination of in vitro and in vivo based assays, we establish a mechanism for cystine recognition and proton coupled transport. Mutational mapping and functional characterisation of human cystinosin further provide a framework for understanding the molecular impact of disease-causing mutations.
- Department of Biochemistry, University of Oxford, Oxford, UK.
Organizational Affiliation: