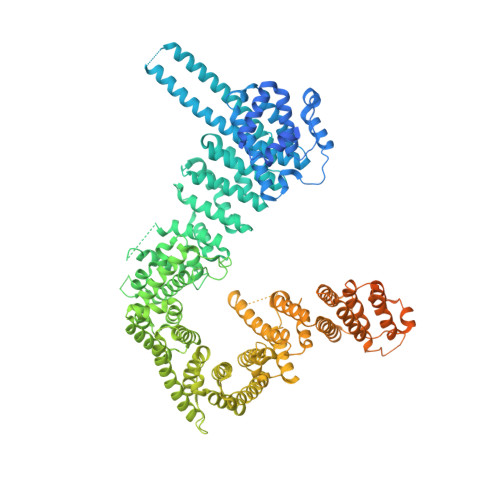
Structural basis of centromeric cohesion protection.
Garcia-Nieto, A., Patel, A., Li, Y., Oldenkamp, R., Feletto, L., Graham, J.J., Willems, L., Muir, K.W., Panne, D., Rowland, B.D.(2023) Nat Struct Mol Biol 30: 853-859
- PubMed: 37081319
- DOI: https://doi.org/10.1038/s41594-023-00968-y
- Primary Citation of Related Structures:
7ZJS - PubMed Abstract:
In the early stages of mitosis, cohesin is released from chromosome arms but not from centromeres. The protection of centromeric cohesin by SGO1 maintains the sister chromatid cohesion that resists the pulling forces of microtubules until all chromosomes are attached in a bipolar manner to the mitotic spindle. Here we present the X-ray crystal structure of a segment of human SGO1 bound to a conserved surface of the cohesin complex. SGO1 binds to a composite interface formed by the SA2 and SCC1 RAD21 subunits of cohesin. SGO1 shares this binding interface with CTCF, indicating that these distinct chromosomal regulators control cohesin through a universal principle. This interaction is essential for the localization of SGO1 to centromeres and protects centromeric cohesin against WAPL-mediated cohesin release. SGO1-cohesin binding is maintained until the formation of microtubule-kinetochore attachments and is required for faithful chromosome segregation and the maintenance of a stable karyotype.
- Division of Cell Biology, The Netherlands Cancer Institute, Amsterdam, the Netherlands.
Organizational Affiliation: