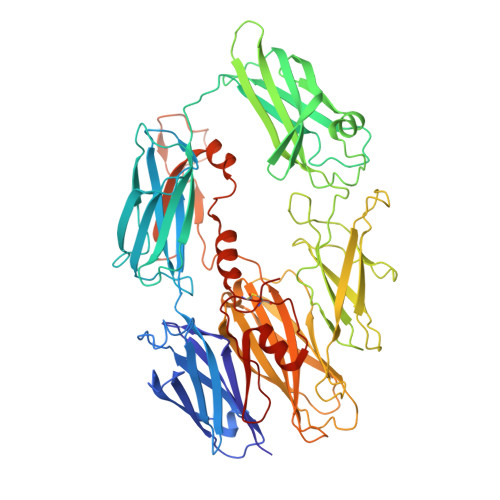
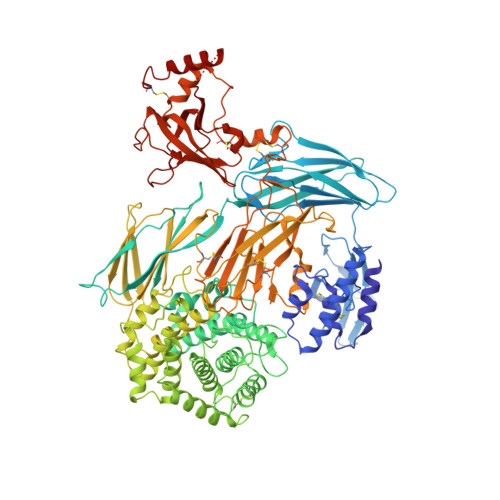
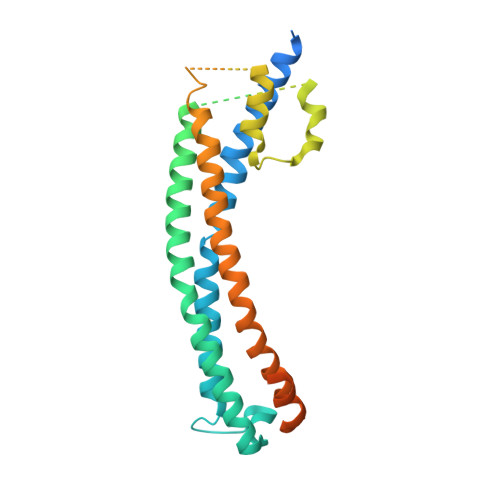
Cryo-EM structures of Trypanosoma brucei gambiense ISG65 with human complement C3 and C3b and their roles in alternative pathway restriction.
Sulzen, H., Began, J., Dhillon, A., Kereiche, S., Pompach, P., Votrubova, J., Zahedifard, F., Subrtova, A., Safner, M., Hubalek, M., Thompson, M., Zoltner, M., Zoll, S.(2023) Nat Commun 14: 2403-2403
- PubMed: 37105991
- DOI: https://doi.org/10.1038/s41467-023-37988-7
- Primary Citation of Related Structures:
7ZGJ, 7ZGK - PubMed Abstract:
African Trypanosomes have developed elaborate mechanisms to escape the adaptive immune response, but little is known about complement evasion particularly at the early stage of infection. Here we show that ISG65 of the human-infective parasite Trypanosoma brucei gambiense is a receptor for human complement factor C3 and its activation fragments and that it takes over a role in selective inhibition of the alternative pathway C5 convertase and thus abrogation of the terminal pathway. No deposition of C4b, as part of the classical and lectin pathway convertases, was detected on trypanosomes. We present the cryo-electron microscopy (EM) structures of native C3 and C3b in complex with ISG65 which reveal a set of modes of complement interaction. Based on these findings, we propose a model for receptor-ligand interactions as they occur at the plasma membrane of blood-stage trypanosomes and may facilitate innate immune escape of the parasite.
- Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo namesti 542/2, 16000, Prague, Czech Republic.
Organizational Affiliation: