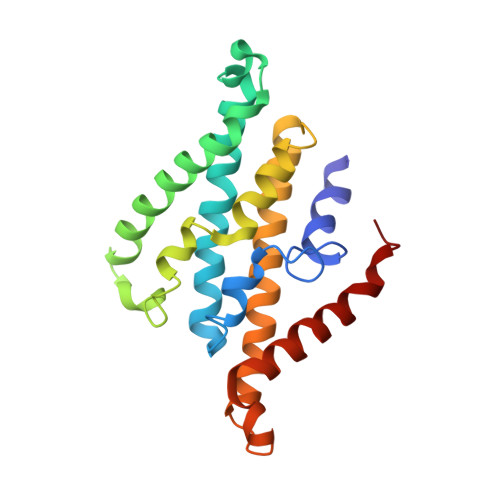
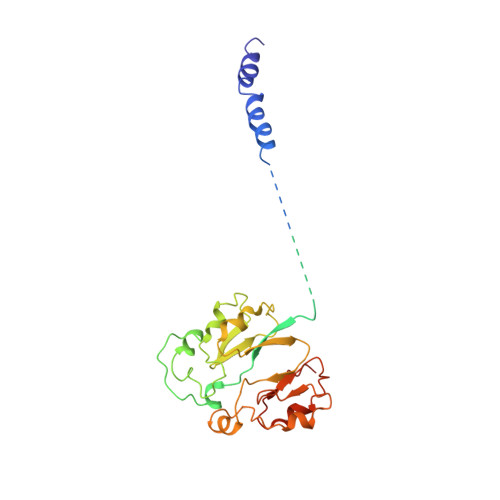
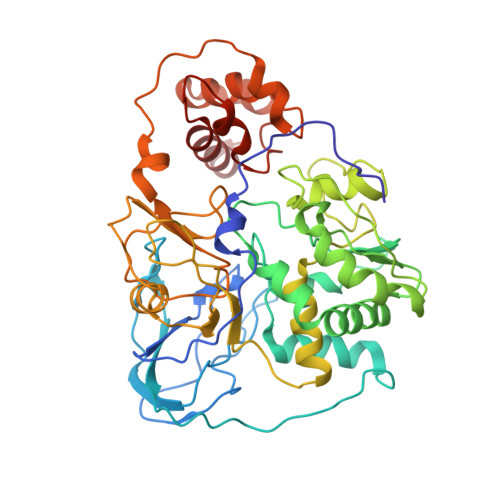
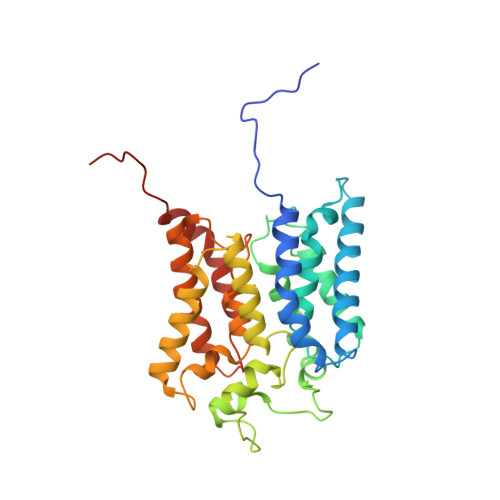
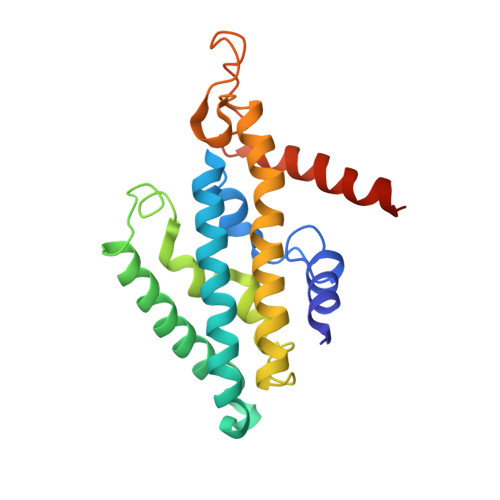
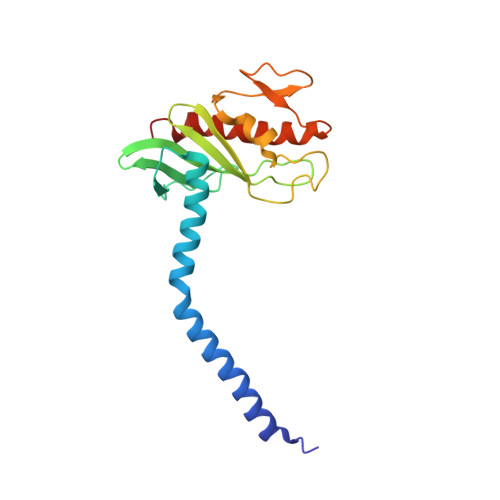
Purification and structural characterization of the Na + -translocating ferredoxin: NAD + reductase (Rnf) complex of Clostridium tetanomorphum.
Vitt, S., Prinz, S., Eisinger, M., Ermler, U., Buckel, W.(2022) Nat Commun 13: 6315-6315
- PubMed: 36274063
- DOI: https://doi.org/10.1038/s41467-022-34007-z
- Primary Citation of Related Structures:
7ZC6 - PubMed Abstract:
Various microbial metabolisms use H + /Na + -translocating ferredoxin:NAD + reductase (Rnf) either to exergonically oxidize reduced ferredoxin by NAD + for generating a transmembrane electrochemical potential or reversely to exploit the latter for producing reduced ferredoxin. For cryo-EM structural analysis, we elaborated a quick four-step purification protocol for the Rnf complex from Clostridium tetanomorphum and integrated the homogeneous and active enzyme into a nanodisc. The obtained 4.27 Å density map largely allows chain tracing and redox cofactor identification complemented by biochemical data from entire Rnf and single subunits RnfB, RnfC and RnfG. On this basis, we postulated an electron transfer route between ferredoxin and NAD via eight [4Fe-4S] clusters, one Fe ion and four flavins crossing the cell membrane twice related to the pathway of NADH:ubiquinone reductase. Redox-coupled Na + translocation is provided by orchestrating Na + uptake/release, electrostatic effects of the assumed membrane-integrated FMN semiquinone anion and accompanied polypeptide rearrangements mediated by different redox steps.
- Department of Molecular Membrane Biology, Max Planck Institute of Biophysics, Max-von-Laue-Str. 3, Frankfurt am Main, Germany. Stella.vitt@mpibp-frankfurt.mpg.de.
Organizational Affiliation: