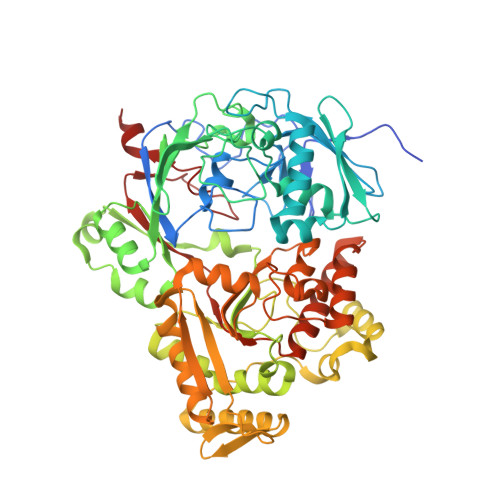
Dual-Uptake Mode of the Antibiotic Phazolicin Prevents Resistance Acquisition by Gram-Negative Bacteria.
Travin, D.Y., Jouan, R., Vigouroux, A., Inaba-Inoue, S., Lachat, J., Haq, F., Timchenko, T., Sutormin, D., Dubiley, S., Beis, K., Morera, S., Severinov, K., Mergaert, P.(2023) mBio 14: e0021723-e0021723
- PubMed: 36802165
- DOI: https://doi.org/10.1128/mbio.00217-23
- Primary Citation of Related Structures:
7Z8E - PubMed Abstract:
Phazolicin (PHZ) is a peptide antibiotic exhibiting narrow-spectrum activity against rhizobia closely related to its producer, Rhizobium sp. strain Pop5. Here, we show that the frequency of spontaneous PHZ-resistant mutants in Sinorhizobium meliloti is below the detection limit. We find that PHZ can enter S. meliloti cells through two distinct promiscuous peptide transporters, BacA and YejABEF, which belong to the SLiPT (SbmA-like peptide transporter) and ABC (ATP-binding cassette) transporter families, respectively. The dual-uptake mode explains the lack of observed resistance acquisition because the simultaneous inactivation of both transporters is necessary for resistance to PHZ. Since both BacA and YejABEF are essential for the development of functional symbiosis of S. meliloti with leguminous plants, the unlikely acquisition of PHZ resistance via the inactivation of these transporters is further disfavored. A whole-genome transposon sequencing screen did not reveal additional genes that can provide strong PHZ resistance when inactivated. However, it was found that the capsular polysaccharide KPS, the novel putative envelope polysaccharide PPP (PHZ-protecting polysaccharide), as well as the peptidoglycan layer jointly contribute to the sensitivity of S. meliloti to PHZ, most likely serving as barriers that reduce the amount of PHZ transported inside the cell. IMPORTANCE Many bacteria produce antimicrobial peptides to eliminate competitors and create an exclusive niche. These peptides act either by membrane disruption or by inhibiting essential intracellular processes. The Achilles' heel of the latter type of antimicrobials is their dependence on transporters to enter susceptible cells. Transporter inactivation results in resistance. Here, we show that a rhizobial ribosome-targeting peptide, phazolicin (PHZ), uses two different transporters, BacA and YejABEF, to enter the cells of a symbiotic bacterium, Sinorhizobium meliloti. This dual-entry mode dramatically reduces the probability of the appearance of PHZ-resistant mutants. Since these transporters are also crucial for S. meliloti symbiotic associations with host plants, their inactivation in natural settings is strongly disfavored, making PHZ an attractive lead for the development of biocontrol agents for agriculture.
- Center of Life Sciences, Skolkovo Institute of Science and Technology, Moscow, Russia.
Organizational Affiliation: