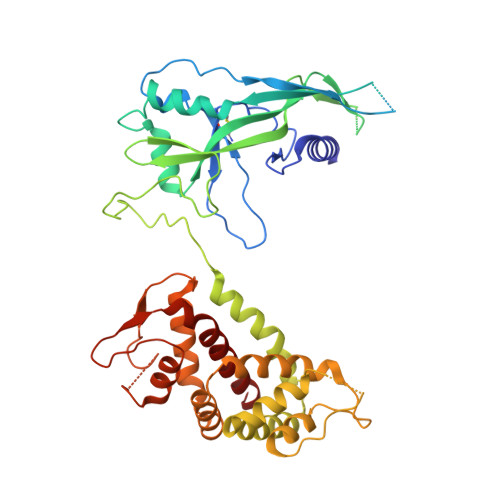
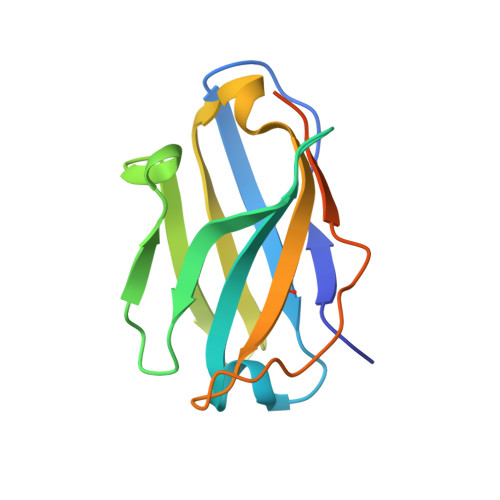
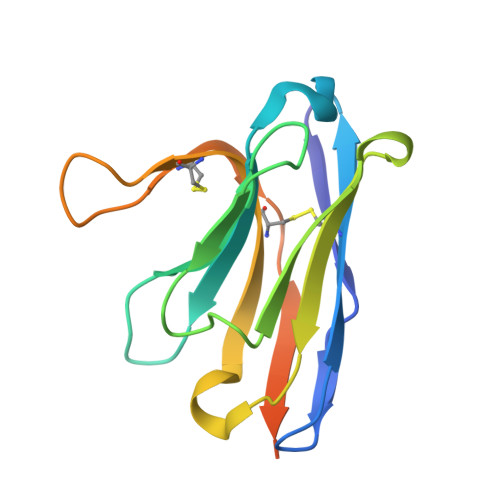
Pyroptosis inhibiting nanobodies block Gasdermin D pore formation.
Kopp, A., Hagelueken, G., Jamitzky, I., Moecking, J., Schiffelers, L.D.J., Schmidt, F.I., Geyer, M.(2023) Nat Commun 14: 7923-7923
- PubMed: 38040708
- DOI: https://doi.org/10.1038/s41467-023-43707-z
- Primary Citation of Related Structures:
7Z1X - PubMed Abstract:
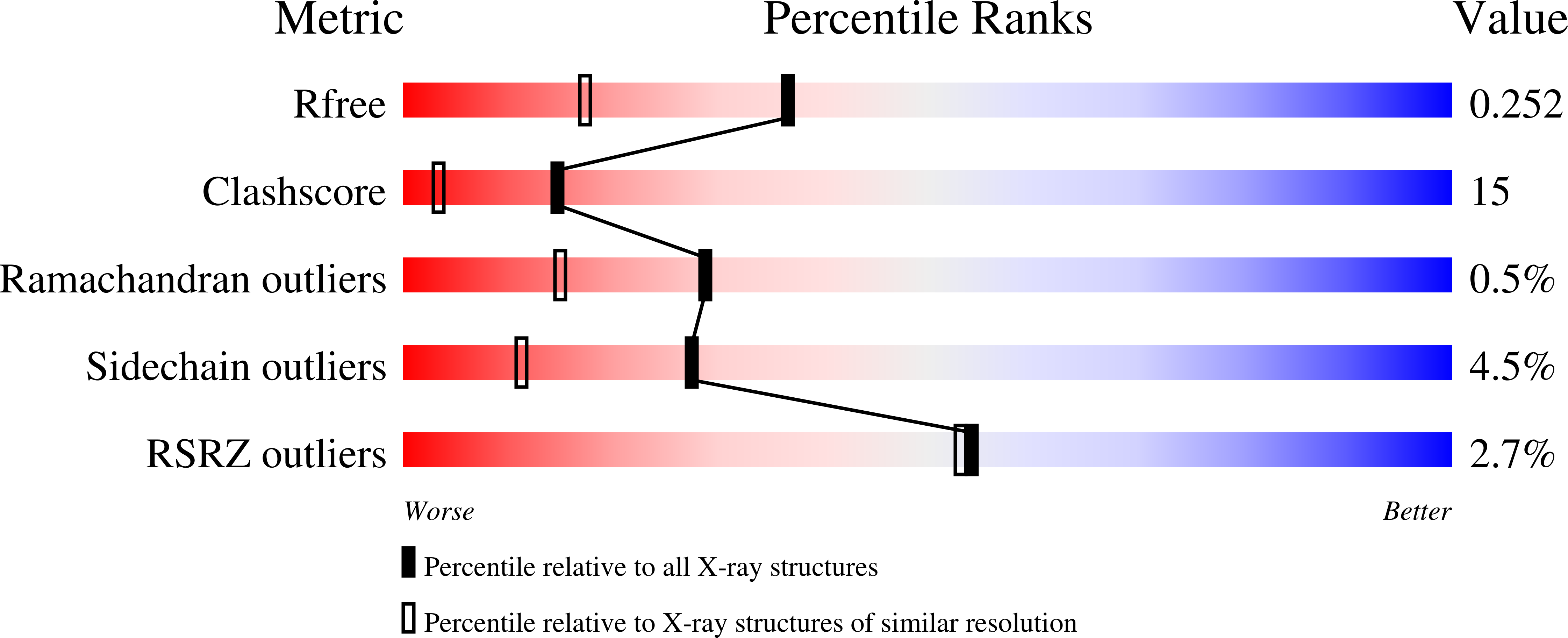
Human Gasdermin D (GSDMD) is a key mediator of pyroptosis, a pro-inflammatory form of cell death occurring downstream of inflammasome activation as part of the innate immune defence. Upon cleavage by inflammatory caspases in the cytosol, the N-terminal domain of GSDMD forms pores in the plasma membrane resulting in cytokine release and eventually cell death. Targeting GSDMD is an attractive way to dampen inflammation. In this study, six GSDMD targeting nanobodies are characterized in terms of their binding affinity, stability, and effect on GSDMD pore formation. Three of the nanobodies inhibit GSDMD pore formation in a liposome leakage assay, although caspase cleavage was not perturbed. We determine the crystal structure of human GSDMD in complex with two nanobodies at 1.9 Å resolution, providing detailed insights into the GSDMD-nanobody interactions and epitope binding. The pore formation is sterically blocked by one of the nanobodies that binds to the oligomerization interface of the N-terminal domain in the multi-subunit pore assembly. Our biochemical and structural findings provide tools for studying inflammasome biology and build a framework for the design of GSDMD targeting drugs.
Organizational Affiliation:
Institute of Structural Biology, Medical Faculty, University of Bonn, Venusberg-Campus 1, 53127, Bonn, Germany.