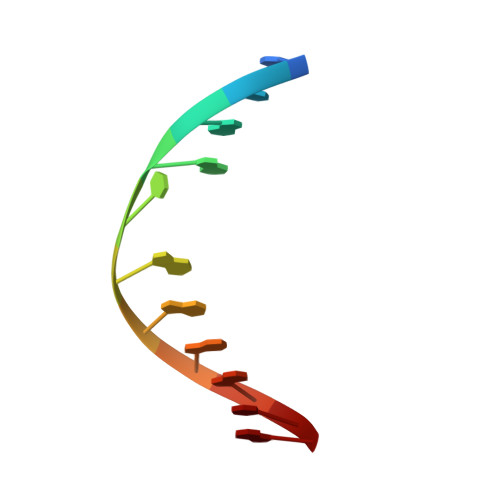
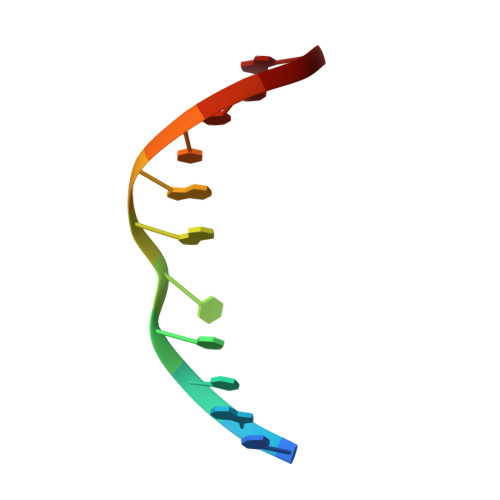
NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.
Yamada, T., Furuita, K., Sakurabayashi, S., Nomura, M., Kojima, C., Nakatani, K.(2022) Nucleic Acids Res 50: 9621-9631
- PubMed: 36095126
- DOI: https://doi.org/10.1093/nar/gkac740
- Primary Citation of Related Structures:
7YVW - PubMed Abstract:
Trinucleotide repeat (TNR) diseases are caused by the aberrant expansion of CXG (X = C, A, G and T) sequences in genomes. We have reported two small molecules binding to TNR, NCD, and NA, which strongly bind to CGG repeat (responsible sequence of fragile X syndrome) and CAG repeat (Huntington's disease). The NMR structure of NA binding to the CAG/CAG triad has been clarified, but the structure of NCD bound to the CGG/CGG triad remained to be addressed. We here report the structural determination of the NCD-CGG/CGG complex by NMR spectroscopy and the comparison with the NA-CAG/CAG complex. While the NCD-CGG/CGG structure shares the binding characteristics with that of the NA-CAG/CAG complex, a significant difference was found in the overall structure caused by the structural fluctuation at the ligand-bound site. The NCD-CGG/CGG complex was suggested in the equilibrium between stacked and kinked structures, although NA-CAG/CAG complex has only the stacked structures. The dynamic fluctuation of the NCD-CGG/CGG structure at the NCD-binding site suggested room for optimization in the linker structure of NCD to gain improved affinity to the CGG/CGG triad.
- Department of Regulatory Bioorganic Chemistry, SANKEN, Osaka University, 8-1 Mihogaoka, Ibaraki 567-0047, Japan.
Organizational Affiliation: