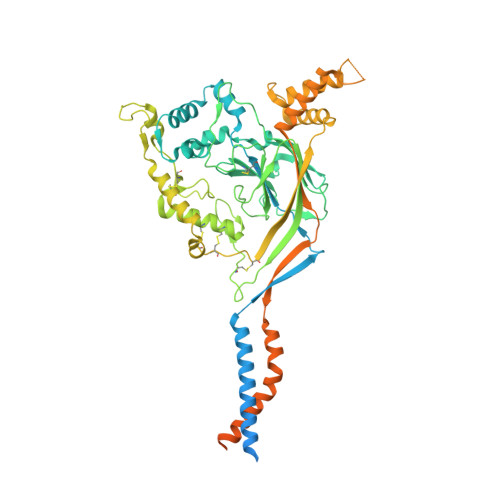
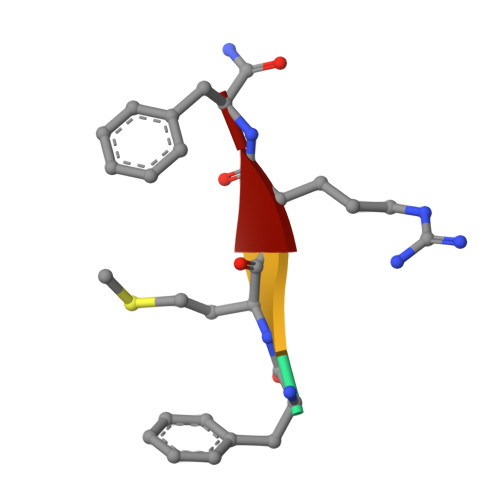
Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Liu, F., Dang, Y., Li, L., Feng, H., Li, J., Wang, H., Zhang, X., Zhang, Z., Ye, S., Tian, Y., Chen, Q.(2023) Nat Chem Biol 19: 1276-1285
- PubMed: 37550431
- DOI: https://doi.org/10.1038/s41589-023-01401-7
- Primary Citation of Related Structures:
7YVB, 7YVC - PubMed Abstract:
Phe-Met-Arg-Phe-amide (FMRFamide)-activated sodium channels (FaNaCs) are a family of channels activated by the neuropeptide FMRFamide, and, to date, the underlying ligand gating mechanism remains unknown. Here we present the high-resolution cryo-electron microscopy structures of Aplysia californica FaNaC in both apo and FMRFamide-bound states. AcFaNaC forms a chalice-shaped trimer and possesses several notable features, including two FaNaC-specific insertion regions, a distinct finger domain and non-domain-swapped transmembrane helix 2 in the transmembrane domain (TMD). One FMRFamide binds to each subunit in a cleft located in the top-most region of the extracellular domain, with participation of residues from the neighboring subunit. Bound FMRFamide adopts an extended conformation. FMRFamide binds tightly to A. californica FaNaC in an N terminus-in manner, which causes collapse of the binding cleft and induces large local conformational rearrangements. Such conformational changes are propagated downward toward the TMD via the palm domain, possibly resulting in outward movement of the TMD and dilation of the ion conduction pore.
- State Key Laboratory of Conservation and Utilization of Bio-Resources in Yunnan and Center for Life Sciences, School of Life Sciences, Yunnan University, Kunming, China.
Organizational Affiliation: