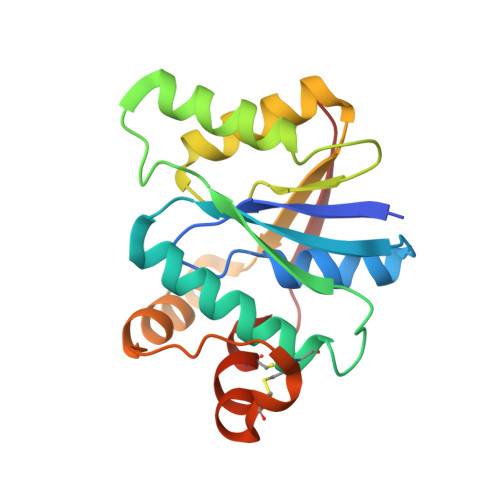
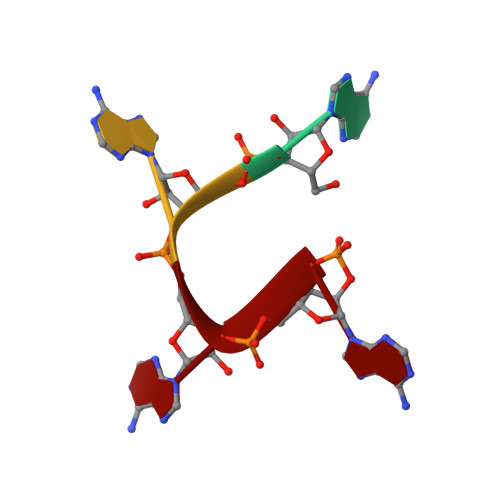
Molecular basis of stepwise cyclic tetra-adenylate cleavage by the type III CRISPR ring nuclease Crn1/Sso2081.
Du, L., Zhang, D., Luo, Z., Lin, Z.(2023) Nucleic Acids Res 51: 2485-2495
- PubMed: 36807980
- DOI: https://doi.org/10.1093/nar/gkad101
- Primary Citation of Related Structures:
7YGH, 7YGL, 7YHL, 8HTW - PubMed Abstract:
The cyclic oligoadenylates (cOAs) act as second messengers of the type III CRISPR immunity system through activating the auxiliary nucleases for indiscriminate RNA degradation. The cOA-degrading nucleases (ring nucleases) provide an 'off-switch' regulation of the signaling, thereby preventing cell dormancy or cell death. Here, we describe the crystal structures of the founding member of CRISPR-associated ring nuclease 1 (Crn1) Sso2081 from Saccharolobus solfataricus, alone, bound to phosphate ions or cA4 in both pre-cleavage and cleavage intermediate states. These structures together with biochemical characterizations establish the molecular basis of cA4 recognition and catalysis by Sso2081. The conformational changes in the C-terminal helical insert upon the binding of phosphate ions or cA4 reveal a gate-locking mechanism for ligand binding. The critical residues and motifs identified in this study provide a new insight to distinguish between cOA-degrading and -nondegrading CARF domain-containing proteins.
- College of Chemistry, Fuzhou University, Fuzhou 350108, China.
Organizational Affiliation: