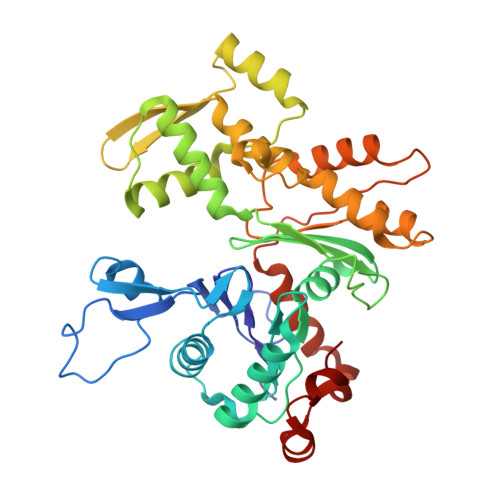
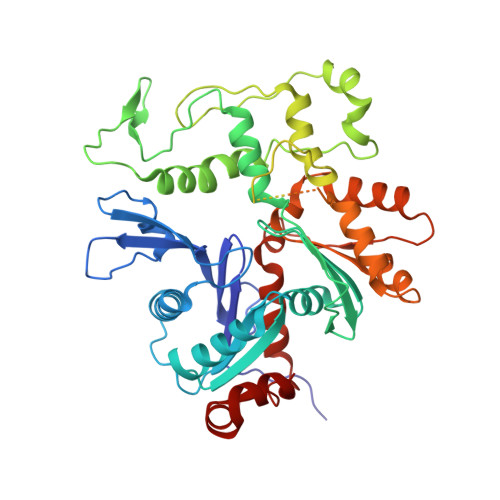
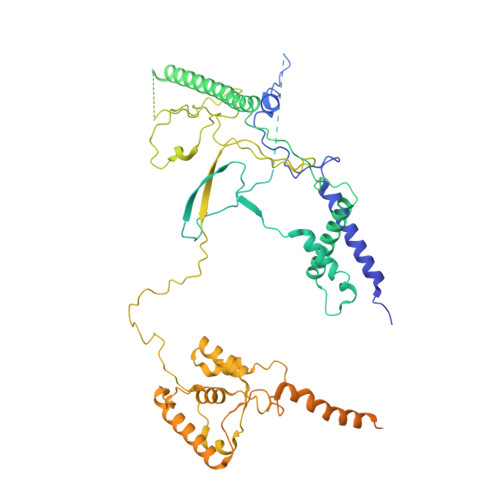
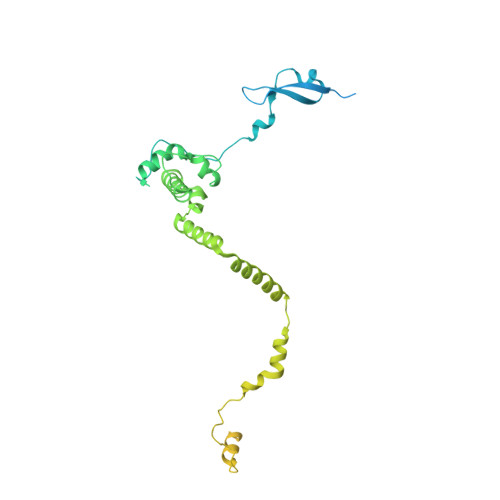
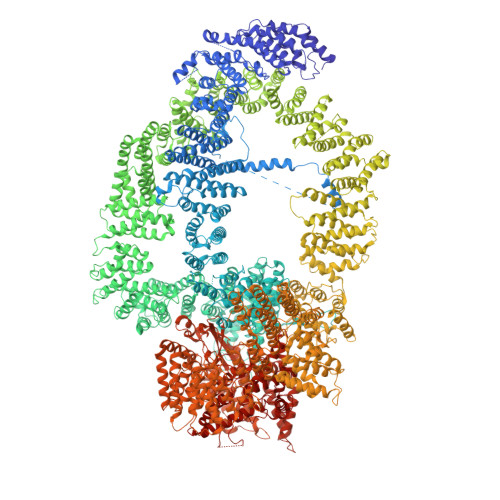
Structure of the NuA4 histone acetyltransferase complex.
Ji, L., Zhao, L., Xu, K., Gao, H., Zhou, Y., Kornberg, R.D., Zhang, H.(2022) Proc Natl Acad Sci U S A 119: e2214313119-e2214313119
- PubMed: 36417436
- DOI: https://doi.org/10.1073/pnas.2214313119
- Primary Citation of Related Structures:
7YFN, 7YFP - PubMed Abstract:
Nucleosome acetyltransferase of H4 (NuA4), one of two major histone acetyltransferase complexes in Saccharomyces cerevisiae specifically acetylates histone H2A and H4, resulting in increased transcriptional activity. Here we present a 3.8-4.0 Å resolution structure of the NuA4 complex from cryoelectron microscopy and associated biochemical studies. The determined structure comprises six subunits and appropriately 5,000 amino acids, with a backbone formed by subunits Eaf1 and Eaf2 spanning from an Actin-Arp4 module to a platform subunit Tra1. Seven subunits are missing from the cryo-EM map. The locations of missing components, Yaf9, and three subunits of the Piccolo module Esa1, Yng2, and Eaf6 were determined. Biochemical studies showed that the Piccolo module and the complete NuA4 exhibit comparable histone acetyltransferase activities, but the Piccolo module binds to nucleosomes, whereas the complete NuA4 does not. The interaction lifetime of NuA4 and nucleosome is evidently short, possibly because of subunits of the NuA4 complex that diminish the affinity of the Piccolo module for the nucleosome, enabling rapid movement from nucleosome to nucleosome.
- Shanghai Institute for Advanced Immunochemical Studies, ShanghaiTech University, Shanghai 201210, China.
Organizational Affiliation: