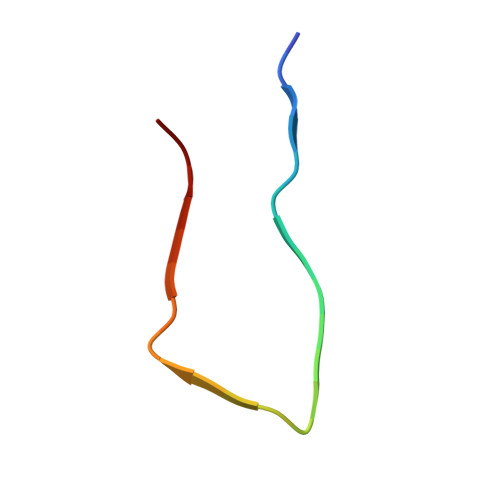
2.2 angstrom Cryo-EM Tetra-Protofilament Structure of the Hamster Prion 108-144 Fibril Reveals an Ordered Water Channel in the Center.
Chen, E.H., Kao, H.W., Lee, C.H., Huang, J.Y.C., Wu, K.P., Chen, R.P.(2022) J Am Chem Soc 144: 13888-13894
- PubMed: 35857020
- DOI: https://doi.org/10.1021/jacs.2c05479
- Primary Citation of Related Structures:
7YAT - PubMed Abstract:
Fibrils of the hamster prion peptide (sHaPrP, sequence 108-144) were prepared in an acidic solution, and their structure was solved by cryogenic electron microscopy with a resolution of 2.23 Å based on the gold-standard Fourier shell correlation (FSC) curve. The fibril has a novel architecture that has never been found in other amyloid fibrils. Each fibril is assembled by four protofilaments (PFs) and has an ordered water channel in the center. Each protofilament contains three β-strands (125-130, 133-135, and 138-141) arranged in an "R"-shaped construct. The structural data indicate that these three β-strand segments are the most amyloidogenic region of the prion peptide/protein and might be the site of nucleation during fibrillization under conditions without denaturants.
- Institute of Biological Chemistry, Academia Sinica, No. 128, Section 2, Academia Road, Nankang, Taipei 115, Taiwan.
Organizational Affiliation: