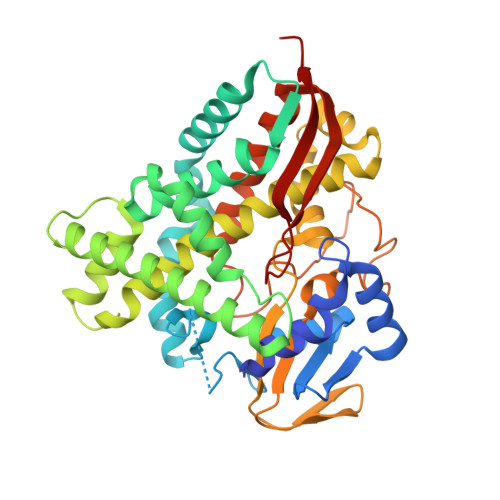
Rationally Controlling Selective Steroid Hydroxylation via Scaffold Sampling of a P450 Family
Zhang, X., Shen, P., Zhao, J., Chen, Y., Li, X., Huang, J.W., Zhang, L., Li, Q., Gao, C., Xing, Q., Chen, C.C., Guo, R.T., Li, A.(2023) ACS Catal 13: 1280-1289