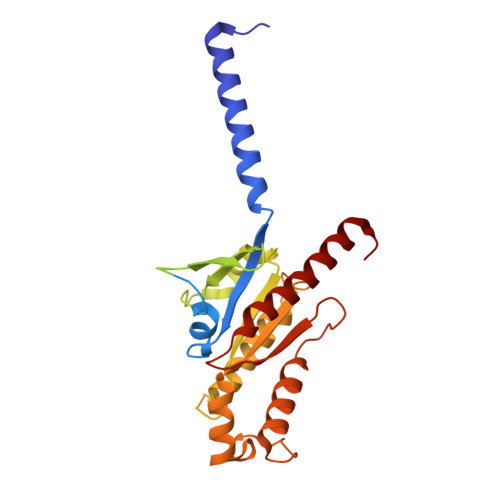
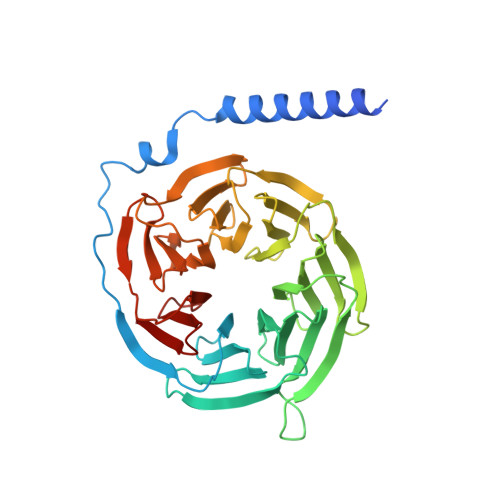
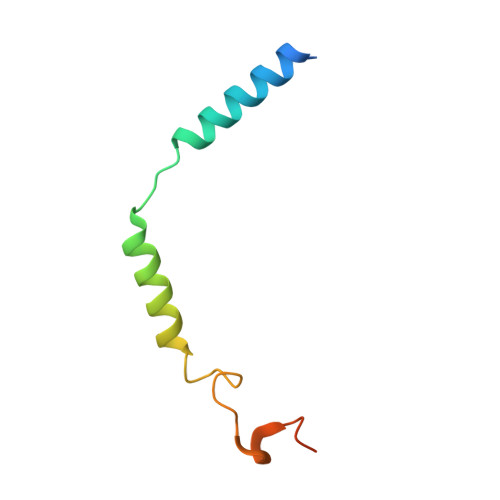
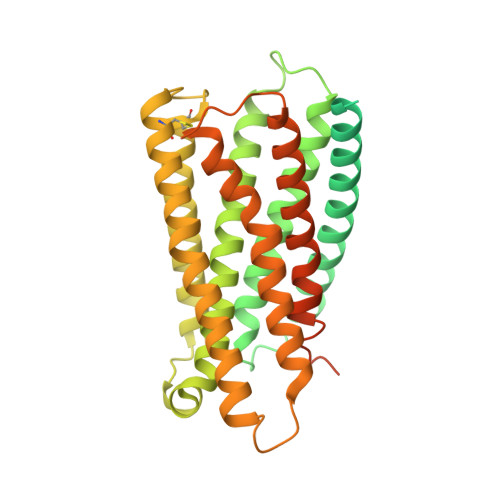
Structural insight into the activation mechanism of MrgD with heterotrimeric Gi-protein revealed by cryo-EM.
Suzuki, S., Iida, M., Hiroaki, Y., Tanaka, K., Kawamoto, A., Kato, T., Oshima, A.(2022) Commun Biol 5: 707-707
- PubMed: 35840655
- DOI: https://doi.org/10.1038/s42003-022-03668-3
- Primary Citation of Related Structures:
7Y12, 7Y13, 7Y14, 7Y15 - PubMed Abstract:
MrgD, a member of the Mas-related G protein-coupled receptor (MRGPR) family, has high basal activity for Gi activation. It recognizes endogenous ligands, such as β-alanine, and is involved in pain and itch signaling. The lack of a high-resolution structure for MrgD hinders our understanding of whether its activation is ligand-dependent or constitutive. Here, we report two cryo-EM structures of the MrgD-Gi complex in the β-alanine-bound and apo states at 3.1 Å and 2.8 Å resolution, respectively. These structures show that β-alanine is bound to a shallow pocket at the extracellular domains. The extracellular half of the sixth transmembrane helix undergoes a significant movement and is tightly packed into the third transmembrane helix through hydrophobic residues, creating the active form. Our structures demonstrate a structural basis for the characteristic ligand recognition of MrgD. These findings provide a framework to guide drug designs targeting the MrgD receptor.
- Department of Basic Medicinal Sciences, Graduate School of Pharmaceutical Sciences, Nagoya University, Furo-cho, Chikusa-ku, Nagoya, 464-8601, Japan.
Organizational Affiliation: