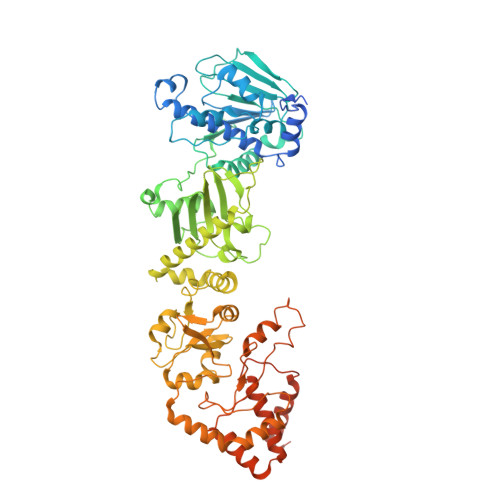
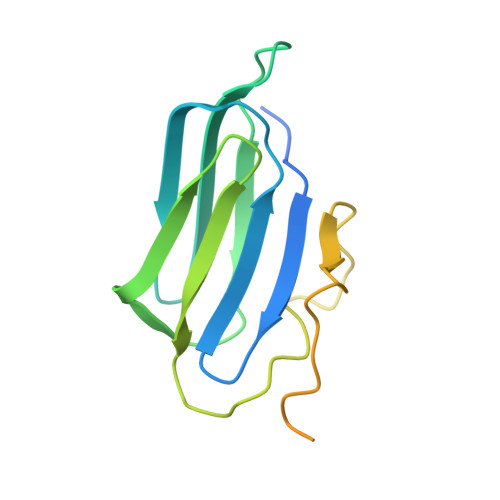
Cryo-EM structure of the cytosolic AhR complex.
Wen, Z., Zhang, Y., Zhang, B., Hang, Y., Xu, L., Chen, Y., Xie, Q., Zhao, Q., Zhang, L., Li, G., Zhao, B., Sun, F., Zhai, Y., Zhu, Y.(2023) Structure 31: 295
- PubMed: 36649707
- DOI: https://doi.org/10.1016/j.str.2022.12.013
- Primary Citation of Related Structures:
7Y04, 8H77 - PubMed Abstract:
Aryl hydrocarbon receptor (AhR) is an important ligand-activated transcription factor involved in the regulation of various important physiological functions. Here, we report the cryo-EM structures of the Hsp90-AhR-p23 complex with or without bound XAP2, where the structure of the mouse AhR PAS-B domain is resolved. A highly conserved bridge motif of AhR is responsible for the interaction with the Hsp90 dimeric lumen. The ligand-free AhR PAS-B domain is attached to the Hsp90 dimer and is stabilized in the complex with bound XAP2. In addition, the DE-loop and a group of conserved pocket inner residues in the AhR PAS-B domain are found to be important for ligand binding. These results reveal the structural basis of the biological functions of AhR. Moreover, the protein purification method presented here allows the isolation of stable mouse AhR protein, which could be used to develop high-sensitivity biosensors for environmental pollutant detection.
- National Key Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: