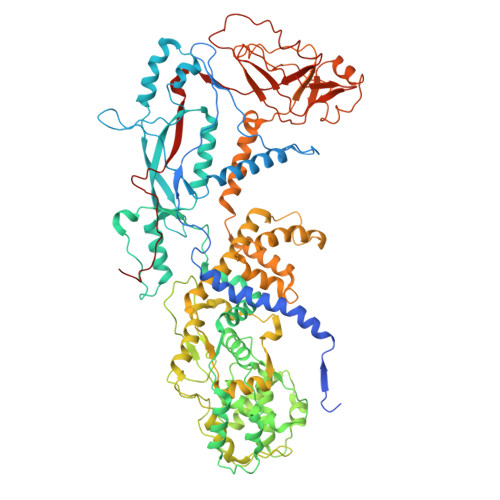
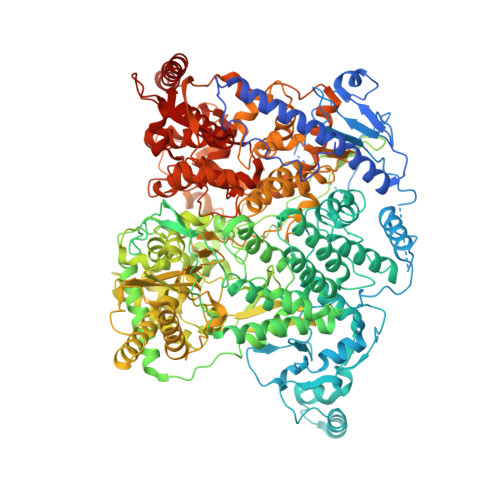
The structure of a 12-segmented dsRNA reovirus: New insights into capsid stabilization and organization.
Zhang, Q., Gao, Y., Baker, M.L., Liu, S., Jia, X., Xu, H., He, J., Kaelber, J.T., Weng, S., Jiang, W.(2023) PLoS Pathog 19: e1011341-e1011341
- PubMed: 37083840
- DOI: https://doi.org/10.1371/journal.ppat.1011341
- Primary Citation of Related Structures:
7XR2, 7XR3 - PubMed Abstract:
Infecting a wide range of hosts, members of Reovirales (formerly Reoviridae) consist of a genome with different numbers of segmented double stranded RNAs (dsRNA) encapsulated by a proteinaceous shell and carry out genome replication and transcription inside the virion. Several cryo-electron microscopy (cryo-EM) structures of reoviruses with 9, 10 or 11 segmented dsRNA genomes have revealed insights into genome arrangement and transcription. However, the structure and genome arrangement of 12-segmented Reovirales members remain poorly understood. Using cryo-EM, we determined the structure of mud crab reovirus (MCRV), a 12-segmented dsRNA virus that is a putative member of Reovirales in the non-turreted Sedoreoviridae family, to near-atomic resolutions with icosahedral symmetry (3.1 Å) and without imposing icosahedral symmetry (3.4 Å). These structures revealed the organization of the major capsid proteins in two layers: an outer T = 13 layer consisting of VP12 trimers and unique VP11 clamps, and an inner T = 1 layer consisting of VP3 dimers. Additionally, ten RNA dependent RNA polymerases (RdRp) were well resolved just below the VP3 layer but were offset from the 5-fold axes and arranged with D5 symmetry, which has not previously been seen in other members of Reovirales. The N-termini of VP3 were shown to adopt four unique conformations; two of which anchor the RdRps, while the other two conformations are likely involved in genome organization and capsid stability. Taken together, these structures provide a new level of understanding for capsid stabilization and genome organization of segmented dsRNA viruses.
- State key lab for biocontrol, School of Life Sciences, Sun Yat-sen University, Guangzhou, China.
Organizational Affiliation: