Crystal structure of T2R-TTL-15 complex
Lun, T., Chengyong, W.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin alpha-1B chain | 450 | Sus scrofa | Mutation(s): 0 Gene Names: TUBA1B EC: 3.6.5 | 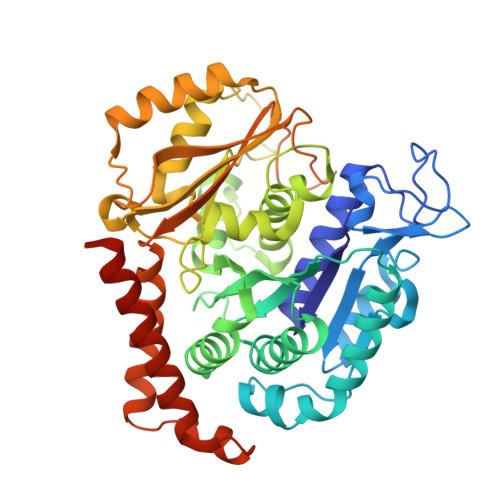 | |
UniProt | |||||
Find proteins for Q2XVP4 (Sus scrofa) Explore Q2XVP4 Go to UniProtKB: Q2XVP4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2XVP4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Tubulin beta chain | 445 | Sus scrofa | Mutation(s): 0 Gene Names: LOC100624785 | 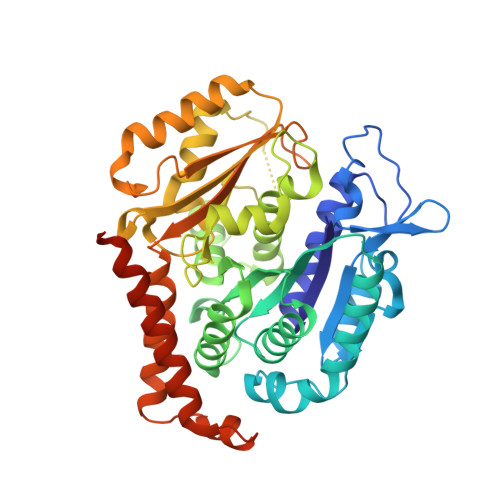 | |
UniProt | |||||
Find proteins for P02554 (Sus scrofa) Explore P02554 Go to UniProtKB: P02554 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02554 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Stathmin-4 | 143 | Mus musculus | Mutation(s): 0 Gene Names: Stmn4 | 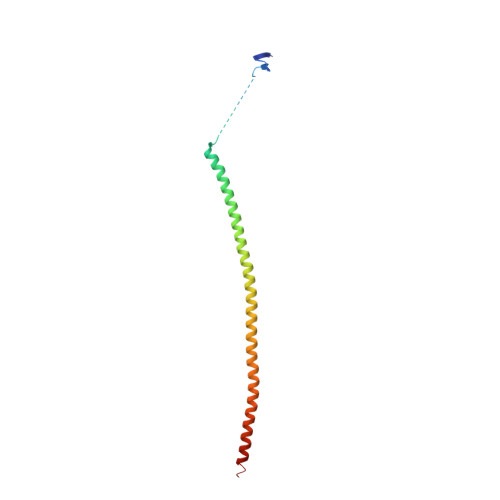 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P63042 (Mus musculus) Explore P63042 Go to UniProtKB: P63042 | |||||
IMPC: MGI:1931224 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P63042 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| TTL | 384 | Gallus gallus | Mutation(s): 0 Gene Names: TTL | 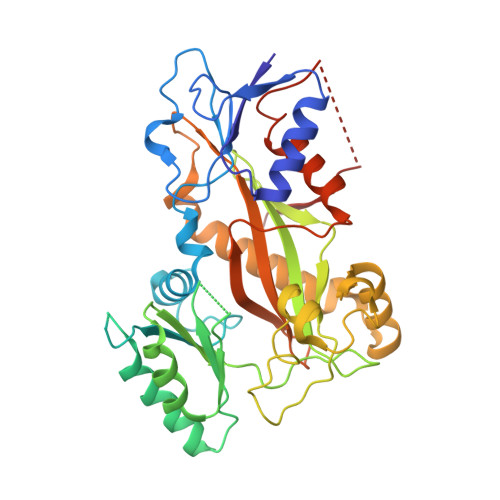 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GTP Query on GTP | G [auth A], P [auth C] | GUANOSINE-5'-TRIPHOSPHATE C10 H16 N5 O14 P3 XKMLYUALXHKNFT-UUOKFMHZSA-N |  | ||
| GDP Query on GDP | K [auth B], S [auth D] | GUANOSINE-5'-DIPHOSPHATE C10 H15 N5 O11 P2 QGWNDRXFNXRZMB-UUOKFMHZSA-N |  | ||
| GX5 (Subject of Investigation/LOI) Query on GX5 | O [auth B], T [auth D] | 2-chloranyl-N-(4-methoxyphenyl)-N-methyl-pyrido[3,2-d]pyrimidin-4-amine C15 H13 Cl N4 O RMEGSJKWVTVCMY-UHFFFAOYSA-N |  | ||
| MES Query on MES | M [auth B], N [auth B] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| CA Query on CA | I [auth A], R [auth C] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | J [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| MG Query on MG | H [auth A], L [auth B], Q [auth C] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 105.42 | α = 90 |
| b = 158.33 | β = 90 |
| c = 180.98 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Aimless | data scaling |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Not funded | -- |