Methylation guide RNAs without box C/D motifs.
Wang, J., Yang, Z., Ye, K.(2022) RNA 28: 1597-1605
- PubMed: 36127125
- DOI: https://doi.org/10.1261/rna.079379.122
- Primary Citation of Related Structures:
7XPL - PubMed Abstract:
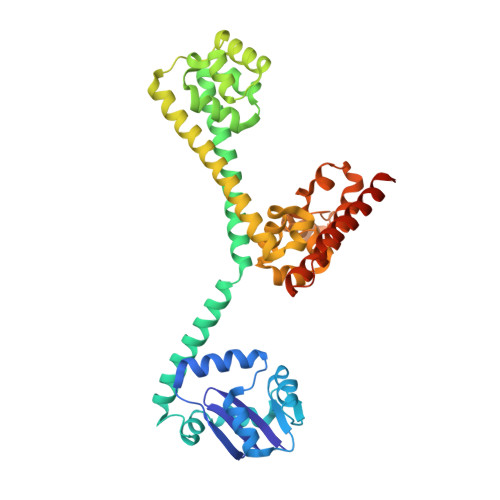
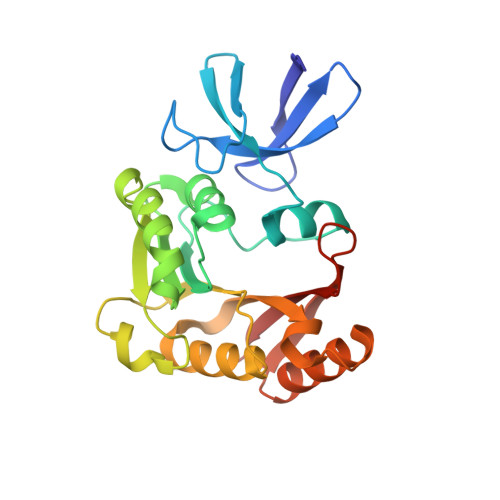
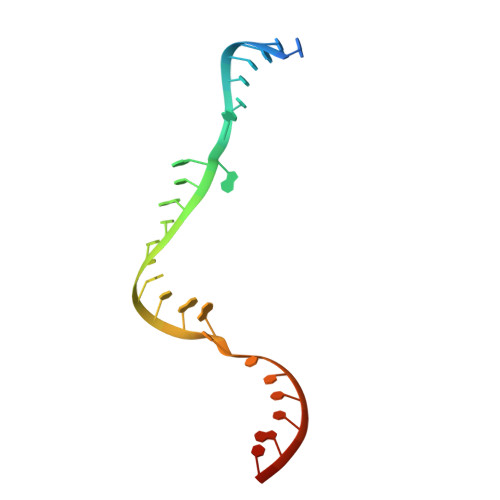
Box C/D RNAs guide site-specific 2'-O-methylation of RNAs in archaea and eukaryotes. The defining feature of methylation guide RNAs is two sets of box C and D motifs that form kink-turn structures specifically recognized by L7Ae family proteins. Here, we engineered a new type of methylation guide that lacks C/D motifs and requires no L7Ae for assembly and function. We determined a crystal structure of a bipartite C/D-free guide RNA in complex with Nop5, fibrillarin and substrate in the active form at 2.2 Å resolution. The stems of new guide RNAs functionally replace C/D motifs in Nop5 binding, precisely placing the substrate for site-specific modification. We also found that the bipartite architecture and association of L7Ae with C/D motifs enhance modification when association of guide RNAs or substrates is weak. Our study provides insights into the variations, robustness and possible evolutionary path of methylation guide RNAs.
- Key Laboratory of RNA Biology, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: