Blockade of dual immune checkpoint inhibitory signals with a CD47/PD-L1 bispecific antibody for cancer treatment.
Wang, R., Zhang, C., Cao, Y., Wang, J., Jiao, S., Zhang, J., Wang, M., Tang, P., Ouyang, Z., Liang, W., Mao, Y., Wang, A., Li, G., Zhang, J., Wang, M., Wang, S., Gui, X.(2023) Theranostics 13: 148-160
- PubMed: 36593962
- DOI: https://doi.org/10.7150/thno.79367
- Primary Citation of Related Structures:
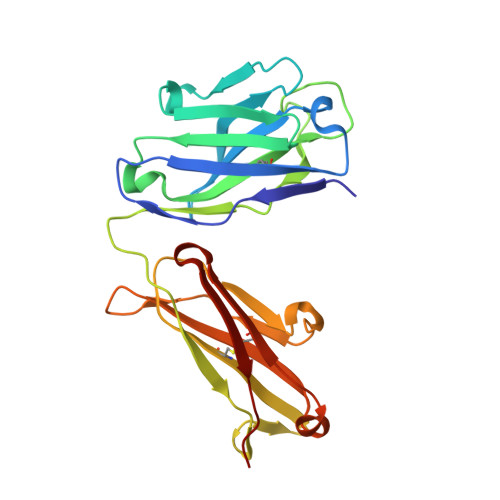
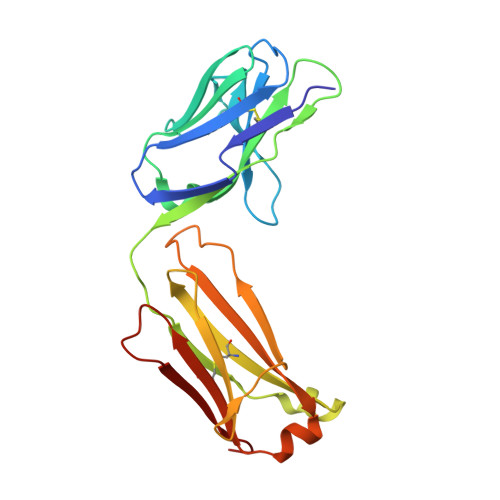
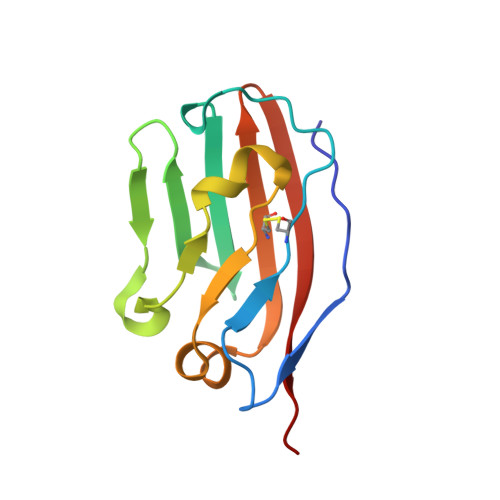
7XJF - PubMed Abstract:
Background: Even though PD-1/PD-L1 is an identified key "don't find me" signal to active adaptive immune system for cancer treatment, the overall response rate (ORR) for all cancer patients is still limited. Other effective therapeutic modalities to bridge the innate and adaptive immunity to improve ORR are urgently needed. Recently, CD47/SIRPα interaction is confirmed as a critical "don't eat me" signal to active innate immunity. However, the red blood cell (RBC) toxicity is the big concern for the development of CD47-based anti-cancer therapeutics. Methods: Here, we report the development of a CD47/PD-L1 bispecific antibody 6MW3211 to block both PD-1/PD-L1 and CD47/SIRPα signals, and studied the effects of 6MW3211 on anti-tumor immune functions in vitro and in vivo . The pharmacokinetic and toxicity profiles of 6MW3211 were evaluated in GLP non-human primate (NHP) studies. Results: The dual immune checkpoint inhibitory signaling blocker 6MW3211 shows high binding affinity to PD-L1 and low binding affinity to CD47. This inequivalent binding affinity design makes 6MW3211 preferentially bound to PD-L1 on tumor cells followed by disrupting the interaction of CD47/SIRPα. Complex structure determination and flow cytometry assay demonstrated that 6MW3211 has no binding to either human or rhesus monkey RBCs. 6MW3211 effectively blocked both PD-1/DP-L1 and CD47/SIRPα signaling and promoted macrophage phagocytosis of tumor cells. Potent therapeutic efficacies of 6MW3211 in three different mouse models were further observed. Moreover, 6MW3211 was demonstrated to have a fairly good safety profile in a GLP NHP study. In addition, multiplex fluorescent immunohistochemistry (mIHC) staining shows that PD-L1 and CD47 co-express on several different types of human tumor tissues. Conclusions: These results support the development of 6MW3211 for the treatment of PD-L1 and CD47 double positive cancers.
- Mabwell (Shanghai) Bioscience Co., Ltd., Shanghai 201210, China.
Organizational Affiliation: