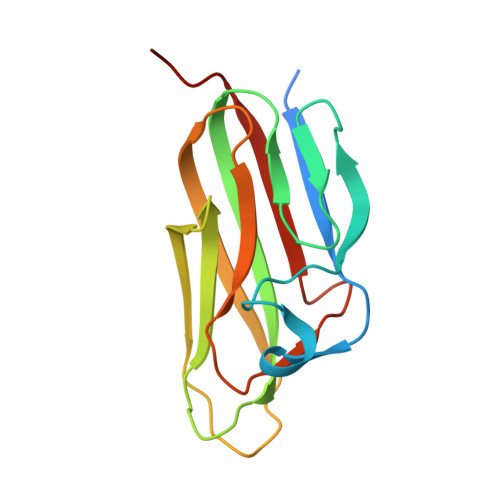
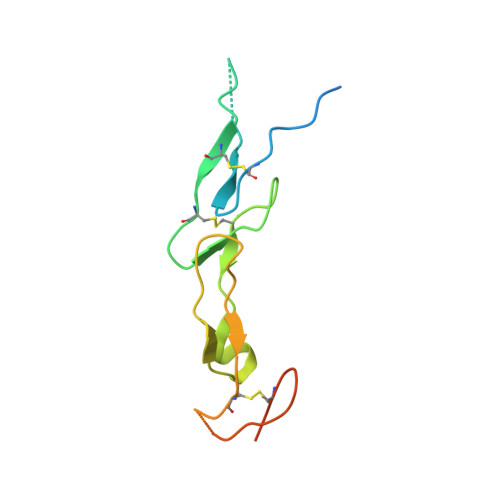
Structural insights into pathogenic mechanism of hypohidrotic ectodermal dysplasia caused by ectodysplasin A variants.
Yu, K., Huang, C., Wan, F., Jiang, C., Chen, J., Li, X., Wang, F., Wu, J., Lei, M., Wu, Y.(2023) Nat Commun 14: 767-767
- PubMed: 36765055
- DOI: https://doi.org/10.1038/s41467-023-36367-6
- Primary Citation of Related Structures:
7X9G - PubMed Abstract:
EDA is a tumor necrosis factor (TNF) family member, which functions together with its cognate receptor EDAR during ectodermal organ development. Mutations of EDA have long been known to cause X-linked hypohidrotic dysplasia in humans characterized by primary defects in teeth, hair and sweat glands. However, the structural information of EDA interaction with EDAR is lacking and the pathogenic mechanism of EDA variants is poorly understood. Here, we report the crystal structure of EDA C-terminal TNF homology domain bound to the N-terminal cysteine-rich domains of EDAR. Together with biochemical, cellular and mouse genetic studies, we show that different EDA mutations lead to varying degrees of ectodermal developmental defects in mice, which is consistent with the clinical observations on human patients. Our work extends the understanding of the EDA signaling mechanism, and provides important insights into the molecular pathogenesis of disease-causing EDA variants.
- Department of Second Dental Center, Ninth People's Hospital Affiliated with Shanghai Jiao Tong University, School of Medicine, Shanghai Key Laboratory of Stomatology, National Clinical Research Center of Stomatology, Shanghai, China.
Organizational Affiliation: