Structural basis of peptidomimetic agonism revealed by small- molecule GLP-1R agonists Boc5 and WB4-24.
Cong, Z., Zhou, Q., Li, Y., Chen, L.N., Zhang, Z.C., Liang, A., Liu, Q., Wu, X., Dai, A., Xia, T., Wu, W., Zhang, Y., Yang, D., Wang, M.W.(2022) Proc Natl Acad Sci U S A 119: e2200155119-e2200155119
- PubMed: 35561211
- DOI: https://doi.org/10.1073/pnas.2200155119
- Primary Citation of Related Structures:
7X8R, 7X8S - PubMed Abstract:
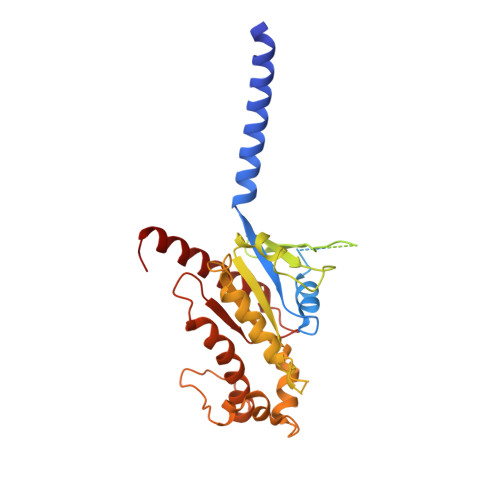
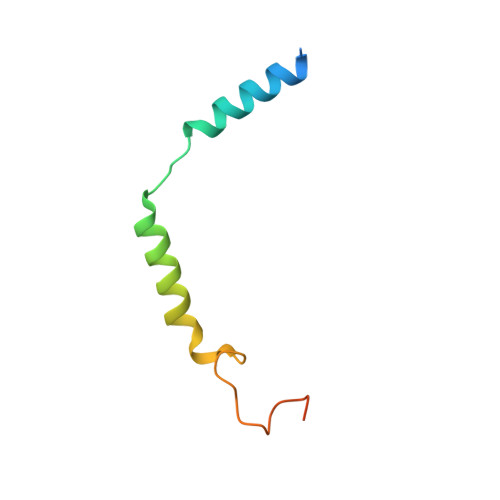
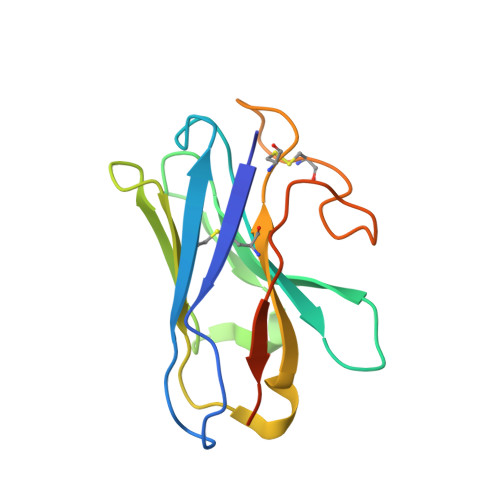
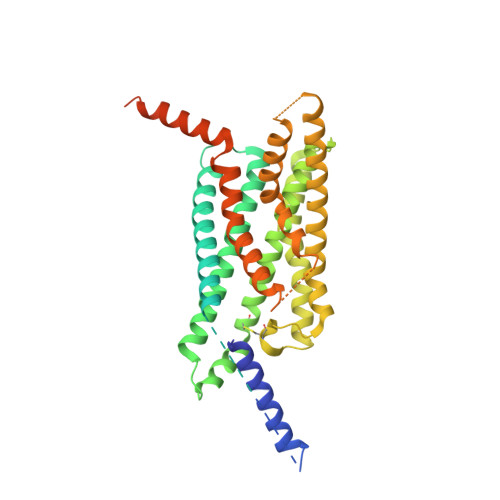
Glucagon-like peptide-1 receptor (GLP-1R) agonists are effective in treating type 2 diabetes and obesity with proven cardiovascular benefits. However, most of these agonists are peptides and require subcutaneous injection except for orally available semaglutide. Boc5 was identified as the first orthosteric nonpeptidic agonist of GLP-1R that mimics a broad spectrum of bioactivities of GLP-1 in vitro and in vivo. Here, we report the cryoelectron microscopy structures of Boc5 and its analog WB4-24 in complex with the human GLP-1R and Gs protein. Bound to the extracellular domain, extracellular loop 2, and transmembrane (TM) helices 1, 2, 3, and 7, one arm of both compounds was inserted deeply into the bottom of the orthosteric binding pocket that is usually accessible by peptidic agonists, thereby partially overlapping with the residues A8 to D15 in GLP-1. The other three arms, meanwhile, extended to the TM1-TM7, TM1-TM2, and TM2-TM3 clefts, showing an interaction feature substantially similar to the previously known small-molecule agonist LY3502970. Such a unique binding mode creates a distinct conformation that confers both peptidomimetic agonism and biased signaling induced by nonpeptidic modulators at GLP-1R. Further, the conformational difference between Boc5 and WB4-24, two closed related compounds, provides a structural framework for fine-tuning of pharmacological efficacy in the development of future small-molecule therapeutics targeting GLP-1R.
- Department of Pharmacology, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Organizational Affiliation: