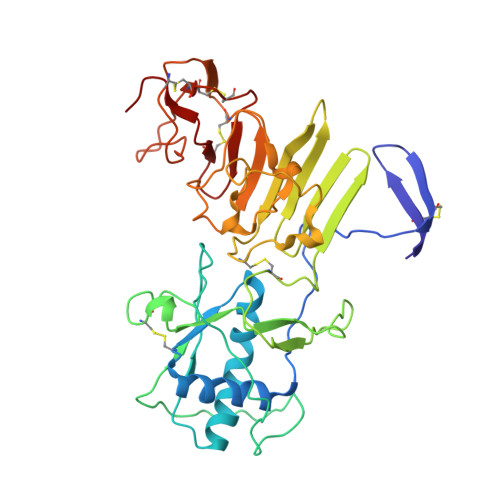
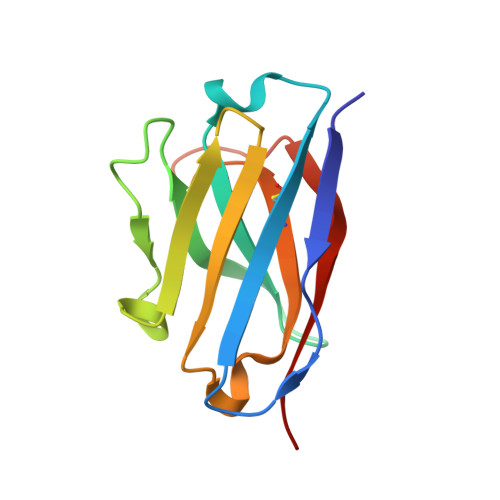
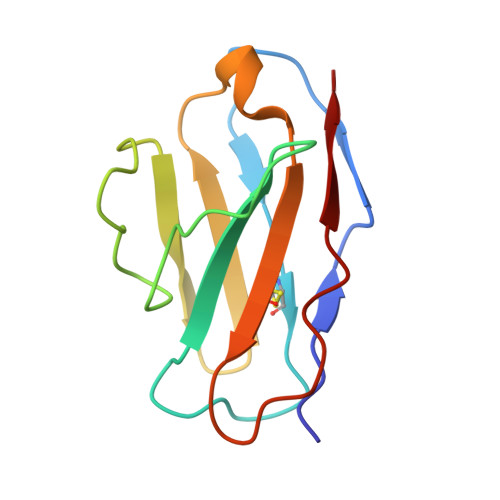
CryoEM structures of the multimeric secreted NS1, a major factor for dengue hemorrhagic fever.
Shu, B., Ooi, J.S.G., Tan, A.W.K., Ng, T.S., Dejnirattisai, W., Mongkolsapaya, J., Fibriansah, G., Shi, J., Kostyuchenko, V.A., Screaton, G.R., Lok, S.M.(2022) Nat Commun 13: 6756-6756
- PubMed: 36347841
- DOI: https://doi.org/10.1038/s41467-022-34415-1
- Primary Citation of Related Structures:
7WUR, 7WUS, 7WUT, 7WUU, 7WUV - PubMed Abstract:
Dengue virus infection can cause dengue hemorrhagic fever (DHF). Dengue NS1 is multifunctional. The intracellular dimeric NS1 (iNS1) forms part of the viral replication complex. Previous studies suggest the extracellular secreted NS1 (sNS1), which is a major factor contributing to DHF, exists as hexamers. The structure of the iNS1 is well-characterised but not that of sNS1. Here we show by cryoEM that the recombinant sNS1 exists in multiple oligomeric states: the tetrameric (stable and loose conformation) and hexameric structures. Stability of the stable and loose tetramers is determined by the conformation of their N-terminal domain - elongated β-sheet or β-roll. Binding of an anti-NS1 Fab breaks the loose tetrameric and hexameric sNS1 into dimers, whereas the stable tetramer remains largely unbound. Our results show detailed quaternary organization of different oligomeric states of sNS1 and will contribute towards the design of dengue therapeutics.
- Programme in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore, 169857, Singapore.
Organizational Affiliation: