Structural and functional insights into a novel pre-clinical-stage antibody targeting IL-17A for treatment of autoimmune diseases.
Xu, J.G., Jia, H., Chen, S., Xu, J., Zhan, Y., Yu, H., Wang, W., Kang, X., Cui, X., Feng, Y., Chen, X., Xu, W., Pan, X., Wei, X., Li, H., Wang, Y., Xia, S., Liu, X., Yang, L., He, Y., Zhu, X.(2022) Int J Biol Macromol 202: 529-538
- PubMed: 35066019
- DOI: https://doi.org/10.1016/j.ijbiomac.2022.01.119
- Primary Citation of Related Structures:
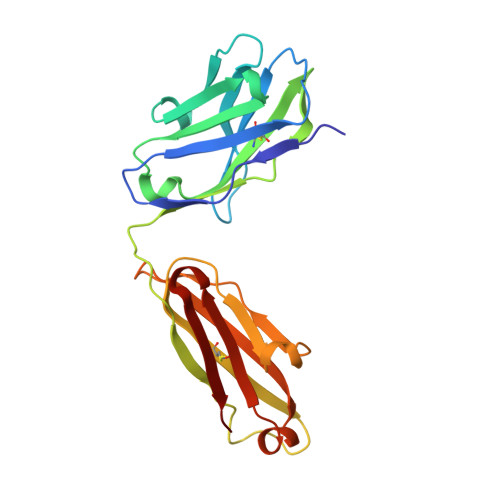
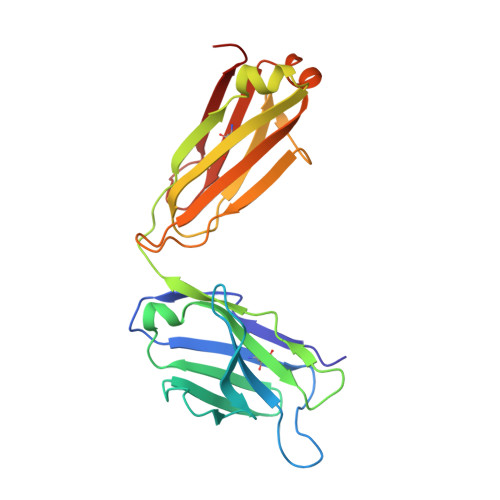
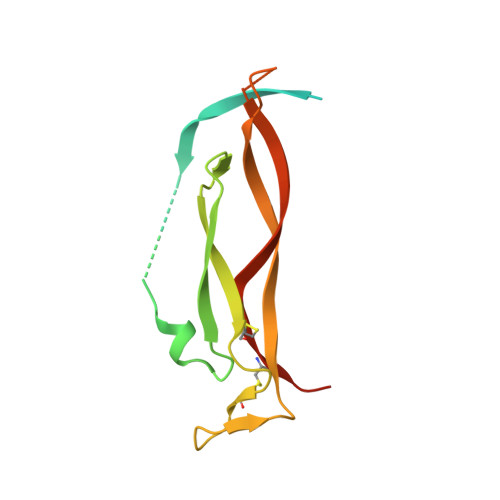
7WKX - PubMed Abstract:
The pro-inflammatory cytokine interleukin-17A (IL-17A) is a key driver of multiple inflammatory and immune disorders. Therapeutic antibodies targeting IL-17A have been proven effective in treating patients with these diseases; however, large variations in clinical outcomes have been observed with different antibodies. In this study, we developed HB0017, a novel monoclonal antibody that targets human IL-17A. HB0017 specifically and strongly bound to human, cynomolgus monkey, and mouse IL-17A at the physiological interface with the IL-17A receptor. In human and monkey cells, HB0017 potently antagonized the functions of IL-17A through competitive binding. HB0017 functioned equivalently to that of clinically approved antibodies in terms of therapeutic efficacy for inflammatory disorders and psoriasis in a mouse model. The results indicate that HB0017 may be an alternative biological therapy for treating patients with inflammation and autoimmune diseases.
- Drug Discovery, Shanghai Huaota Biopharmaceutical Co. Ltd., Shanghai 201203, China.
Organizational Affiliation: