A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
Zhang, J., Lu, D., Li, M., Liu, M., Yao, S., Zhan, J., Liu, W.J., Gao, G.F.(2022) China CDC Wkly 4: 83-87
- PubMed: 35186375
- DOI: https://doi.org/10.46234/ccdcw2021.258
- Primary Citation of Related Structures:
7WKJ - PubMed Abstract:
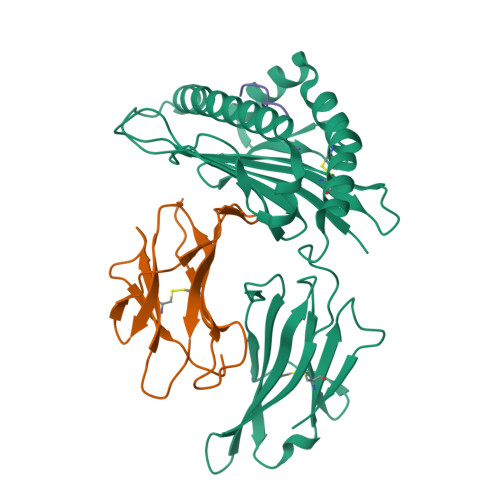
Similar to antibody detection, severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2)-specific T-cell response evaluation is also pivotal among the coronavirus disease 2019 (COVID-19) convalescents and the vaccinated populations. Nucleocapsid (N) protein is one of the main structural proteins of SARS-CoV-2 and can trigger T-cell responses in humans. An overlapping peptide pool covering the full length of the N protein was designed, peptides with positive T-cell activating potency in COVID-19 convalescents were screened, and CD8 + T cell epitopes were further identified. The epitope was used to detect the SARS-CoV-2-specific CD8 + T cell responses in COVID-19 convalescents based in intracellular cytokine staining and tetramer staining in flow cytometry. A human leukocyte antigen A (HLA-A)*1101-restricted CD8 + T cell epitope, which could stimulate the production of IFN-γ via peripheral blood mononuclear cells (PBMCs) of the convalescents was defined, and the tetramer generated with this epitope could detect SARS-CoV-2-specific T cells in the PBMCs of the convalescents. The structural investigation eliminated that the epitope was a typical HLA-A*1101-restricted T-cell epitope which was conserved among all the sarbecoviruses. The newly identified SARS-CoV-2-derived T-cell epitope was helpful to detect the cellular immunity against different sarbecoviruses including SARS-CoV and SARS-CoV-2. This study provided an evaluation method and also a peptide candidate for the research and development of T-cell based vaccine for the virus.
Organizational Affiliation:
NHC Key Laboratory of Biosafety, National Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China.