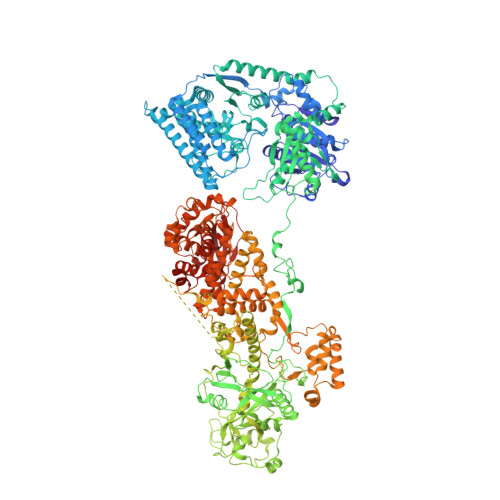
Structural insights into dsRNA processing by Drosophila Dicer-2-Loqs-PD.
Su, S., Wang, J., Deng, T., Yuan, X., He, J., Liu, N., Li, X., Huang, Y., Wang, H.W., Ma, J.(2022) Nature 607: 399-406
- PubMed: 35768513
- DOI: https://doi.org/10.1038/s41586-022-04911-x
- Primary Citation of Related Structures:
7W0A, 7W0B, 7W0C, 7W0D, 7W0E, 7W0F - PubMed Abstract:
Small interfering RNAs (siRNAs) are the key components for RNA interference (RNAi), a conserved RNA-silencing mechanism in many eukaryotes 1,2 . In Drosophila, an RNase III enzyme Dicer-2 (Dcr-2), aided by its cofactor Loquacious-PD (Loqs-PD), has an important role in generating 21 bp siRNA duplexes from long double-stranded RNAs (dsRNAs) 3,4 . ATP hydrolysis by the helicase domain of Dcr-2 is critical to the successful processing of a long dsRNA into consecutive siRNA duplexes 5,6 . Here we report the cryo-electron microscopy structures of Dcr-2-Loqs-PD in the apo state and in multiple states in which it is processing a 50 bp dsRNA substrate. The structures elucidated interactions between Dcr-2 and Loqs-PD, and substantial conformational changes of Dcr-2 during a dsRNA-processing cycle. The N-terminal helicase and domain of unknown function 283 (DUF283) domains undergo conformational changes after initial dsRNA binding, forming an ATP-binding pocket and a 5'-phosphate-binding pocket. The overall conformation of Dcr-2-Loqs-PD is relatively rigid during translocating along the dsRNA in the presence of ATP, whereas the interactions between the DUF283 and RIIIDb domains prevent non-specific cleavage during translocation by blocking the access of dsRNA to the RNase active centre. Additional ATP-dependent conformational changes are required to form an active dicing state and precisely cleave the dsRNA into a 21 bp siRNA duplex as confirmed by the structure in the post-dicing state. Collectively, this study revealed the molecular mechanism for the full cycle of ATP-dependent dsRNA processing by Dcr-2-Loqs-PD.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai, China.