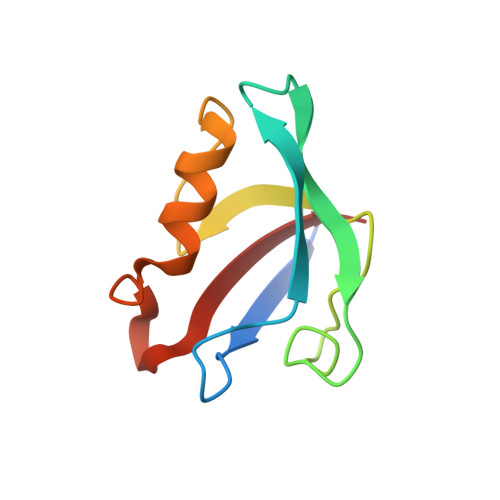
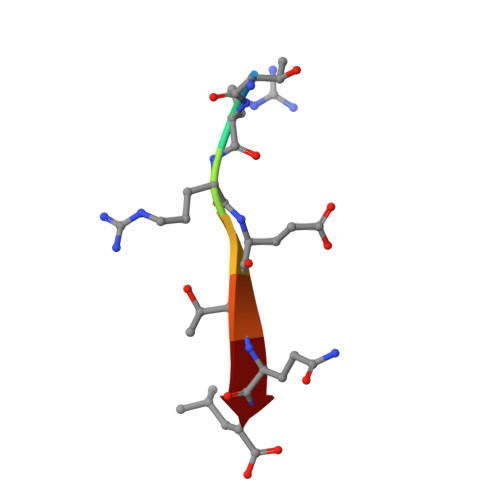
Structural and biochemical analysis of the PTPN4 PDZ domain bound to the C-terminal tail of the human papillomavirus E6 oncoprotein.
Lee, H.S., Yun, H.Y., Lee, E.W., Shin, H.C., Kim, S.J., Ku, B.(2022) J Microbiol 60: 395-401
- PubMed: 35089587
- DOI: https://doi.org/10.1007/s12275-022-1606-1
- Primary Citation of Related Structures:
7VZE - PubMed Abstract:
High-risk genotypes of human papillomaviruses (HPVs) are directly implicated in various abnormalities associated with cellular hyperproliferation, including cervical cancer. E6 is one of two oncoproteins encoded in the HPV genome, which recruits diverse PSD-95/Dlg/ZO-1 (PDZ) domain-containing human proteins through its C-terminal PDZ-binding motif (PBM) to be degraded by means of the proteasome pathway. Among the three PDZ domain-containing protein tyrosine phosphatases, protein tyrosine phosphatase non-receptor type 3 (PTPN3) and PTPN13 were identified to be recognized by HPV E6 in a PBM-dependent manner. However, whether HPV E6 associates with PTPN4, which also has a PDZ domain and functions as an apoptosis regulator, remains undetermined. Herein, we present structural and biochemical evidence demonstrating the direct interaction between the PBM of HPV16 E6 and the PDZ domain of human PTPN4 for the first time. X-ray crystallographic structure determination and binding measurements using isothermal titration calorimetry demonstrated that hydrophobic interactions in which Leu158 of HPV16 E6 plays a key role and a network of intermolecular hydrogen bonds sustain the complex formation between PTPN4 PDZ and the PBM of HPV16 E6. In addition, it was verified that the corresponding motifs from several other high-risk HPV genotypes, including HPV18, HPV31, HPV33, and HPV45, bind to PTPN4 PDZ with comparable affinities, suggesting that PTPN4 is a common target of various pathogenic HPV genotypes.
- Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, 34141, Republic of Korea.
Organizational Affiliation: