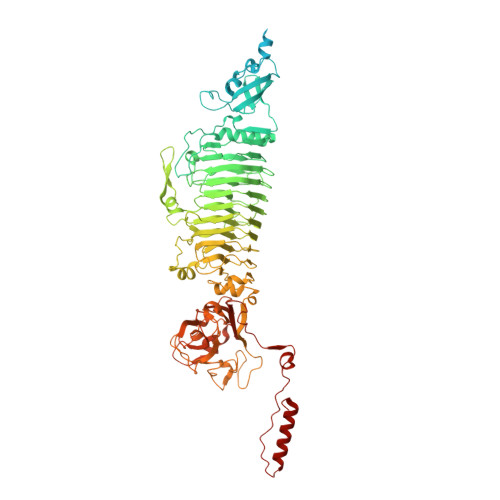
Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Cai, R., Ren, Z., Zhao, R., Lu, Y., Wang, X., Guo, Z., Song, J., Xiang, W., Du, R., Zhang, X., Han, W., Ru, H., Gu, J.(2023) Microbiol Spectr 11: e0530422-e0530422
- PubMed: 37750730
- DOI: https://doi.org/10.1128/spectrum.05304-22
- Primary Citation of Related Structures:
7VYV, 7VZ3 - PubMed Abstract:
Hypervirulent Klebsiella pneumoniae with capsular polysaccharides (CPSs) causes severe nosocomial- and community-acquired infections. Phage-derived depolymerases can degrade CPSs from K. pneumoniae to attenuate bacterial virulence, but their antimicrobial mechanisms and clinical potential are not well understood. In the present study, Klebsiella phage GH-K3-derived depolymerase Depo32 (encoded by gene gp32 ) was identified to exhibit high efficiency in specifically degrading the CPSs of K2 serotype K. pneumoniae . The cryo-electron microscopy structure of trimeric Depo32 at a resolution up to 2.32 Å revealed potential catalytic centers in the cleft of each of the two adjacent subunits. K. pneumoniae subjected to Depo32 became more sensitive to phagocytosis by RAW264.7 cells and activated the cells by the mitogen-activated protein kinase signaling pathway. In addition, intranasal inoculation with Depo32 (a single dose of 200 µg, 20 µg daily for 3 days, or in combination with gentamicin) rescued all C57BL/6J mice infected with a lethal dose of K. pneumoniae K7 without interference from its neutralizing antibody. In summary, this work elaborates on the mechanism by which Depo32 targets the degradation of K2 serotype CPSs and its potential as an antivirulence agent. IMPORTANCE Depolymerases specific to more than 20 serotypes of Klebsiella spp. have been identified, but most studies only evaluated the single-dose treatment of depolymerases with relatively simple clinical evaluation indices and did not reveal the anti-infection mechanism of these depolymerases in depth. On the basis of determining the biological characteristics, the structure of Depo32 was analyzed by cryo-electron microscopy, and the potential active center was further identified. In addition, the effects of Depo32 on macrophage phagocytosis, signaling pathway activation, and serum killing were revealed, and the efficacy of the depolymerase (single treatment, multiple treatments, or in combination with gentamicin) against acute pneumonia caused by Klebsiella pneumoniae was evaluated. Moreover, the roles of the active sites of Depo32 were also elucidated in the in vitro and in vivo studies. Therefore, through structural biology, cell biology, and in vivo experiments, this study demonstrated the mechanism by which Depo32 targets K2 serotype K . pneumoniae infection.
- College of Animal Science and Technology, Jilin Agricultural University, Changchun, Jilin, China.
Organizational Affiliation: